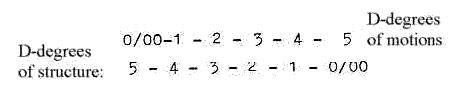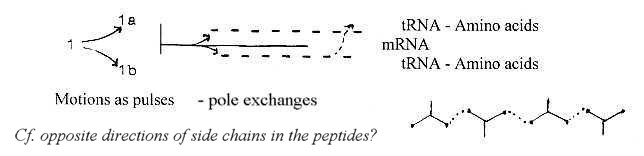|
From DNA to protein chains - some general aspects
At this high, very complicated level there are some remarkable simple
features, which seem to illustrate a dimension chain of the kind
suggested in the pages about
physics:
Dimensional development from D5 to D"0/00"
in the model:
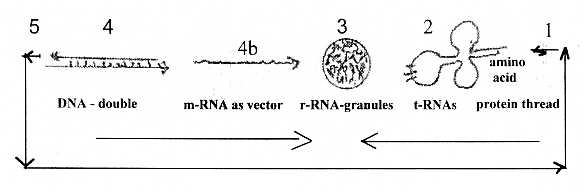
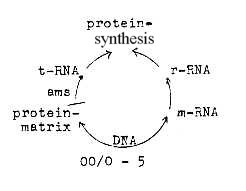
There are about 5 steps in the process:
from DNA→ mRNA → rRNA →tRNA →single amino acid →
peptide chains.
Dimensional geometrical forms in the protein synthesis regarded
on a macro-scale:
Dimension degrees (d-degrees or Dx) shortly:
4 - DNA: double-direction, double strands, opposite directed "vectors"
united,
4b - mRNA, the outward directed"pole" of d-degree 4, with
vector character,
3 - rRNA, the ribosomes, as "balls", volumes,
2 - tRNA, as "plane" forms ("clover leaf" forms)
on a macro-level.
In these forms, it's also possible to see the one
of higher d-degree as binding force in relation to the one of lower
degree in accordance with the model: mRNA binding (or attracting)
ribosomes, ribosomes binding tRNAs, tRNAs binding their special
amino acid.
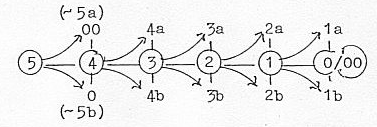
The loop model of a dimension chain, where debranched
degrees in first outward steps "meet the other way around":
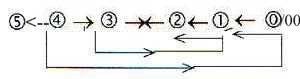
The 5-dimensional chain could be interpreted as double-directed
from, 5→4→3
→etc. and "the other way around"
from 5 to 0/00 and inwards
D4. We have the double DNA-spiral in
the central nucleus of the cell, a double-directed chain, as a vectors
of opposite directions, primarily in the model defined as outward/inward
directions of d-degree 4, with bonds between complementary code
bases A--T, G--C. These pairs may on this underlying level be regarded
as expressions for the outer poles "0 - 00" in d-degree
4. (About the complementary character see here.)
D4b -3:The double chain gets "polarized",
the chains separate - and one gets copied through counterdirection
from the manifold of single code bases in the surrounding (as anticentre")
to mRNA, pole 4b of polarized d-degree 4. As a single stranded
vector it acts like an outward directed force when moving outwards
to the cytoplasm to the ribosomes located between the "centre
and the anticenter" of the cell, between the nucleus and cell
wall.
Cutting of the mRNA, implying loops, may on this
macro-level be regarded as an geometrical illustration of the step
4 → 3 with partly circular forms.
D3-2: rRNA, the ribosomes, have
the 3-dimensional form of small spherical balls where the mRNA gets
attached, however also divided in two parts as in a step 3 --2.
and consisting of both nucleotides and proteins.

Here the meeting between tRNA and rRNA occurs (at right angles,
of natural reasons, but compare the assumed angle 90° in d-degree
3 in the model).
D2-1: tRNAs, transferRNAs, are usually described
with the form of "cloverleaves", principally as such "plane"
structures, partly loops, partly linear, as 2-1-dimensional forms
on a macro-scale; the wavy form (convex/concave as poles of d-degree
2) are also in the model connected with the lower d-degrees.
These shorter chains of nucleotides, different
for different amino acids, represent a manifold as each lower dimension
degree is a manifold in relation to the higher d-degree. In the
middle of the central loop the anti-codon for a certain amino acid
is located.
The anti-codons of tRNAs represent the complementary
strand to mRNA, the 00-pole with inward direction, meeting here
in the middle step 3→2 of the chain.
D1-0/00:
In the opposite end of the tRNA, at the linear end (bases A-C-C
which is similar in all tRNAs), the individual amino acid gets attached.
Then the similar parts of the amino acids, here
called the "B-chains", get bound through condensation
to protein chains.
In this sketch of the process the amino acids (ams) come perhaps
to represent the first 00-pole, meeting "the other way around",
while nucleotides of RNA represent the 0-pole. DNA-nucleotides +
Amino acids as primary poles of a d-degree 5?
00-pole ~ multitude of separate units
Hypothezised is the start in a "whole" made up of DNA
or RNA nucleotides and amino acids combined. A similar direct combination
is part of one of the many theories among scientists trying to explain
the genetic code. Within the "poles" of this assumed unit,
when polarized, the protein synthesis develops.
Some single amino acids take an essential part
in the synthesis of the codon bases.
Counterdirections in the process and the relation mRNA - tRNA:
The difference between T- and U-bases represent the opposite directions:
T-base the inward direction towards the DNA-strands
when not activated. It's replaced by the U-base when a DNA-strand
is copied to mRNA (outward direction) in the synthesis.
The only difference is a CH3-group in the T-base,
obviously the chemical expression for inward direction. (Cf. hydrophobic
bonds.)
Many bases in tRNA have got this group added too,
as a fundamental chemical sign for "inwards" (+ 14 A).
Coordinate axes of the DNA-spiral:
Counting in the first place on three coordinate axes, the first
main axis may be regarded as the one along the base pairing H-bonds:
inwards toward the H-bonds / outwards at copying. An axis of type
d-degree 4 as from a step 5 →4.
Nucleotide pair, e.g. A-T-bases
P-group—deoxiribose—base/R....H ..|. . R—base—(deoxi)ribose—P-group
----------------------------------------> <-------------------------------------------
↓
<--------------------------------------
outwards towards nucleotides as separate units, e. g. AMP...
[Somewhere in the "potential groups"
in the H-bonds (Löwdin) would perhaps the d-degree
5 be found, a representation of "the whole". (!)
Second axis will be the one along along the strands, representing
a d-degree step
4 →3. Secondary H-bonds along the
individual strands curve the strands into helix structures, as we
in the step 4→ 3 in the model get
a transformation toward circular structures and rotation.
The third axis, as a z-axis, should reasonably be expressed
in the relation between mRNA and tRNAs in step 3 →
2 (or more elementary, perhaps originally in the evolution of the
genetic code?) between RNA-nucleotides and proteins or singular
amino acids. (Examples of close relations: Basic amino acids among
others are His and Arg. His derives from the A-base. Arg's R-group
derives from the G-base. Compare the proteins "histones"
on which the DNA-spiral gets rolled up in steps of storage.)
In addition; the opposite directions of side-chains of the amino
acids in the protein chains could eventually be regarded as a forth
coordinate axis:

rRNA - ribosomes:
More details:
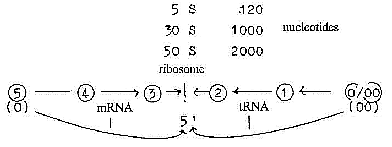
Sketch of a dimensional interpretation:
- We can note the locality of the small "spheres" on
the "endoplasmatic reticulum", a structure of membranes
in cytoplasm - 3-dimensional forms on 2-dimensional surfaces and,
as said above, in "the middle" between the poles centre
- anticentre of the cell.
rRNA is produced in nucleoli, also a spherical organelle.
- It is said that "groups of 5 or 6 individual ribosomes are
held together" by a molecule of mRNA. Why just 5-6? As a number
determined by the dimensions in the model.
It's said too that bacteria has 5 ribosomes! Why
exactly 5?
- We can also note that ribosomes are made up of two parts (50
S and 30 S) with a mass relation 2/1: "50 S" and "30
S": 1,1 x 10E-6 u and 0,55 x 10E-6 u,
- Still another thing about numbers:
Ribosomes consist of about 40 - 60 % RNA, 60 - 40 %
proteins, a 3-2- or 2-3 relation Another reference
says the relation is 64 % RNA, 36 % protein: That would be a relation
82 - 62 : compare "E-numbers" as
sum of poles in d-degrees 3-2 squared, in d-degree 3 = 8, in d-degree
2 = 6.
- How to look at the fact that the ribosomes only act as a kind
of enzymes, a meeting place for mRNA and tRNA and not are involved
in themselves? Doesn't this fact contradicts the general, suggested
interpretation here in terms of the dimension model?
The ribosomes with their close combination of
proteins (amino acids) and bases (nucleotides) seem to illustrate
- on a secondary level - the "whole" hypothezised at start.
The poles from polarized d-degree 5 meet in the middle of the chain.
Hence appear as a secondary evolution of the underlying whole and
as representing a superposed level "detached" to a solely
enzymatic function?
tRNA:
More details.
1. Forms of tRNAs on the macro-scale resemble the 2-dimensional
"clover leafs" with 1-dimensional (partly paired) end
chains and illustrate the geometries convex - concave of poles 2a
/2b suggested in the model.
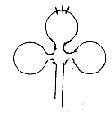
2. tRNAs represent a coordinate axis at straight angle to the the
RNA strand - of natural reasons of course but illustrating the angle
of 90° assumed in d-degree 3.
Further the tRNAs represent a multitude
in relation to the mRNA-chain, a relation connected with poles 00
versus 0.
3. As said above, bases in tRNAs are often methylated, as
the T-base in relation to the U-base, a fact which also points to
the inward direction of tRNAs.
[In the four examples of tRNA given in the used
reference here (Ala, Ser, Phe, Tyr), there is also a T-base in position
23 from the A-C-C-end, a single one. Rather odd. And at this position
appears the same order of the 5 bases: T - U - C - G - A.]
4. The U-base in tRNAs appear in an angled attachment to ribose
as pseudouridine. This fact could be one expression of the
assumed angle steps through the dimension chain. (3 - 2 hypothetically
an angle change from 90° to 45°?)
5. In an alternative form of the dimensional interpretation the
mRNA and tRNAs could be identified with poles of d-degree 4;
polarized to perpendicular relation in d-degree step 3-2: : 4b as
outward direction ~ mRNA, and pole 4a as inward direction ~ tRNAs:
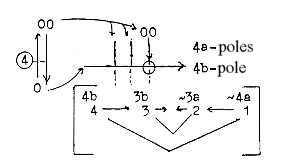
6. With the loop model of the dimension chain the
step 4→3 outwards is also coupled
with step 2←1 inwards.
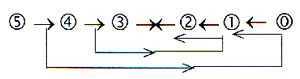
The anti-codons of tRNA corresponds also to the anti-strand of
DNA in relation to mRNA. One could question if the cutting of
mRNA has any inherent connection with tRNAs? Cf. perhaps the
cutting as loop forms and the cleaver leaf form of tRNAs? With its
2-dimensional, partly wavelike form tRNAs have similarities with
the stage where mRNA on its way to rRNA undergoes change: "irrelevant"
(?) parts of the chain are cut off.
|
4 → 3 mRNA
|
|
tRNA 2←1 |
This similarity could be read as a relation between step 4 →
3 outwards and
2←1 inwards, the dimension
chain seen "orthogonal" as double directed with a meeting
in step 3-2.
According to the model we have a 2-dimensional
motion resulting from step 4 → 3,
which could be thought of appearing inwards as a 2-dimensional structure.
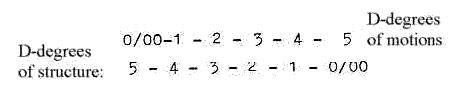
7. Numbers of nucleotides in tRNAs:
It seems to be around 75 - 77: (A little more in tRNA for Ser which
have a beginning of a 4th loop in the leaf. Four examples, counting
from anti-amino acid end to the anti-codon 34 +/- 1 bases, with
anti-codon 37 +/-l.
Including anti-codons:
Ser 85 = 33 + 3 + 49
Tyr 78 = 35 + 3 + 40
Phe 76 = 33 + 3 + 40
Ala 77 = 35 + 3 + 39
One should note that there isn't generally any even number of triplets
in tRNAs, but 26* codons including stop codons times 3 bases give
the number 78. Theoretically one tRNA in a ring form could code
for all amino acids without frame shifting.
*(With the way of counting codons in files about
The genetic code.)
In the 4 examples the quotient G+C /
A+U ≈ 1,4 ≈
45/32.
This relation is the opposite to the quotient
between codons for G+C-coded (10) and A+U-coded (14) amino acids
respectively Same figures in 1st and 2nd base order.) The opposition
- if valid for more tRNAs - could eventually be an expression for
an original opposition in directions between mRNA and tRNAs if the
aspects on codons
on this home page are applied.
8. Why this -C-C-A-end of tRNA, similar in all tRNAs, to
which the amino acids are attached? One aspect concerning mass numbers:
Triplets of a dimension chain inwards and mass
numbers (A) of the bases:
012
111---
123... sum: 135 = A-base
111----
234... sum: 357 = A+C+C.
Sum of the bases with +1 for the bond to ribose.
Intervals between the triplet numbers = 111 = mass number of the
C-base.
345, the last triplet inwards, happens to be the mass-number of
the G-nucleotide in chain binding, also = cGMP.
Compare the triplet series outwards, approximating the sum of side
chains of 20 plus 4 double-coded amino acids:
543, + 1 = G+C-coded ams,
432 + 321 + 210 = 963, - 3 = A+U-coded ams.
Number of bases:
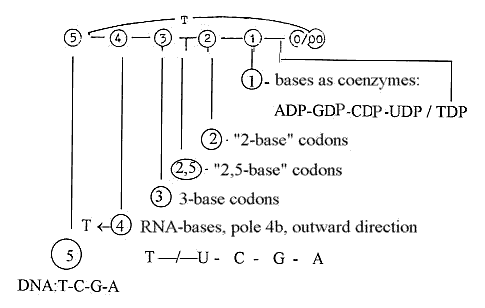
1. 5 bases - 5 dimensions: number 5 returns.
This doesn't mean to say that the bases in any unambiguous sense
represent dimensions or dimension steps. It rather seems as if they
could represent different dimensions in different contexts.
2. 5 bases becomes 4 in DNA, RNA:
T-base in DNA representing direction inwards towards the "mirror",
the H-bridges in the DNA helix. T-base with its CH3-group added
to the U-base made chemically hydrophobic.
U-base in RNA, outward directed towards the protein
synthesis and passing from the nucleus to the outer cell plasma.
Hence, the polarity T/U corresponds to the poles
of d-degree 4, inward / outward directions, DNA←/ → RNA.
3. 4 bases becomes 3 in the codon triplets identifying amino
acids in the protein synthesis. Or 3 - 2: the 3rd base only differentiated
one step in 16 ams, indifferent in 8 ams.
There are other features in the protein synthesis
at rRNA, the ribosomes, which point to the interpretation that this
synthesis occurs in d-degree step 3 - 2.
4. The bases then becomes 1, singular ones, as coenzymes GTP, CTP,
UTP, ATP.
Each dimension step implies one d-degree debranched, transformed
to external motions with the step 1 →
0/00 according to postulates in the model. This means stepwise increasing
motions, revealed in the copying process and protein synthesis -
and the activity of the H-transporting coenzymes.
5. Dimension degree 0/00 (equivalent with 5, as 5') = the d-degree
of Motions, of processes.
Bases as coenzymes as specially connected with elementary kinds
of substances ?
Carbohydrates - Lipids - Proteins - Transportation
T - U
C
G A
T- and U-bases in their role as coenzymes, TTP and
UTP, are used for bonds in carbohydrates (different kinds). The
difference may be seen in UTP(-DP...) active in binding/breaking
of carbohydrates within cells, TTP(-DP...) when it concerns cellulose,
the outer "anticentre" shell of plants.
Note too that all amino acids with U in first
or second position in their codons are derived from stages of the
glycolysis where carbohydrates are broken down.
C as coenzyme CTP is active in synthesis of lipids, in amination
of phospholipids.
G as coenzyme GTP(-DP) takes part in the protein synthesis.
A as coenzyme represent a central energy storage, as such
corresponding to a lot of transportations.
A dimension chain, according to the model here,
implies steps towards growing amount of kinetic energy (motions).
Compare the similar order of the bases in tRNA at position 23...:
T-U-C-G-A!
(If the four examples should be typical.)
Triplets of bases in codons - why?
a) The suggested view could be one elementary answer to the question
of why triplets in the codons. There may be others connected with
the number of coded amino acids.
53 — 43—
33— 233
gives the number chain 125 - 64 - 27 - 8 - 1.
64 theoretically possible different codons becomes
27 in "d-degree" 3, transformed into 19 + 8 (2-base-coded)
ams. 16 ams differentiated in 3rd base + stop codons...(See further
The genetic
code.)
b) Another aspect is that the "loop model" of a dimension
chain implies 3 polarization steps:
5 / 0-00 —>
4 / 1 —> 3 / 2.
(There are also 3 steps in the protein synthesis
from DNA to tRNA:
4 — 3 —
2 — 1: DNA —>
mRNA —> rRNA <—
tRNA.)
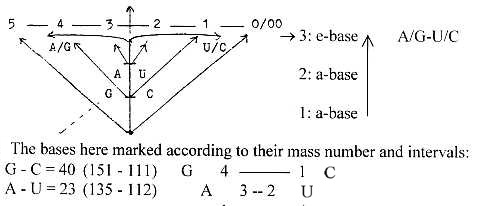
c) Triplets of nucleosides connected by only 2 P-groups?
- Sum of the triplet AUG corresponds to sum of triplets in the elementary
number chain. RNA: Starting code for the protein synthesis = AUG
= amino acid Meth:
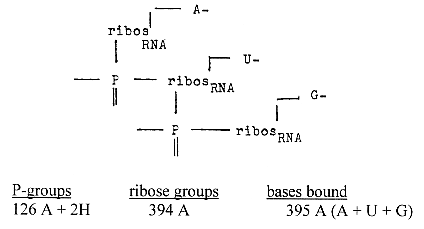
Nucleoside part in chain bonds 2 x 395 -1 = 790
-1 = 792 - 3
Corresponding nucleoside part in DNA with bases T-A-C = 715 = 714
+1
Sum: 1504 equals sum of side-chains of 20 + 4 double-coded
amino acids.
Triplets in the number chain 5 - 4 - 3 - 2 - 1 - 0:
543 345
432 234
321 123
+210 + 012
1506 — 714
Interval 792.
T-A-C-nucleosides: T-A-C-bases bound = 369 = 012+123+234.
Ribose part in DNA: 3(150 - 18 - 16) - 2 H =345 + 1.
d) Numbers from the elementary chain with superposed odd figure
chain:
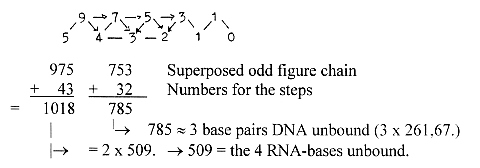
2 x 753 -2 = 1504 = sum of 24 ams R
[A vague association:
The polarization of a line in d-degree 1 into d-degree 0/00 of Motion
has in files about physics been illustrated as in the figure below,
a splitting in steps as virtual lines. This step 1 →
0/00 may be regarded as occurring in each higher steps.
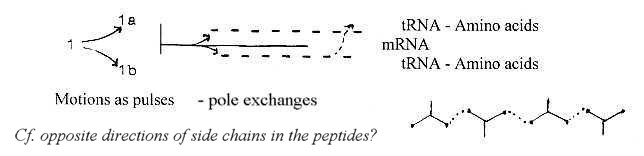
Could we look at the picture as 3 "phases" in the process
of synthesis (?), the balance line + two half steps)
There are also 3 positions A-P-E for a tRNA at
the ribosomes, d-degree 3 in our interpretation, during the process,
the tRNA displaced from A to P to E.
(Groups of 3 half-steps besides the "balance"
line give the picture of B-chains of amino acids with side chains
in opposite directions.)
5-6 as reappearing numbers in steps and storage:
Bonds during the protein synthesis:
5 DNA: H-bridges
4 DNA / mRNA: at copying
3 mRNA / rRNA
2 rRNA / tRNA
1 tRNA / amino acids
00/0 amino acid / amino acid (peptide bonds)
Circa 5 levels of storage of DNA:
Diameter (Ø) = 20 - 100 - 300 - 2000 - 6000 Å):
5 → 4: The double helix of anti-parallel
strands of base-pairs, H-bridge. Ø 20 Å
4 →3: This thread winded up on balls
or cylinders of proteins, (histones), Ø 100 Å
3 →2: This strand of balls drawn
up in a big spiral, Ø 300 Å
2 → 1: This in its turned drawn up
in a thicker spiral, Ø 2000 Å
1 → 0/00: In last step this thicker
spiral drawn up in a still
thicker spiral, Ø 6000 Å, making up the arms of the
X-formed chromosomes, and as a centre of this the centromeres, with
a structure of the type 0/00. Centre - anticenter, ~ 5'.
D-degrees should be viewed as a suggestion. Alternatively we could
see the d-degree of motions, from linear (1), 20 Å, to rotation
(2) on "balls", 100 Å, to winding spirally (3 dimensions),
300 Å, to windings spirally of higher orders in d-degrees
4 to 5, 2000 Å - 6000 Å.
There are 5 histones too, proteins of a certain,
fundamental kind, on which DNA is rolled up: one of these 5 seems
to have a function related with the "all-embracing" character
of "the other way around",

Cf. too 5 bonds between the two pairs of bases:
3 between G- and C-base,
2 between A and U-base.
Helix structures:
H-bonds between protein chains and along the main axis of the chains
are in one reference classified in number of steps:
310-helix: 4,5 steps
α-helix: 3,5
steps
π-helix: 2,5
steps
Protein structures 1 - 2- -3 -4 -:
1 - Primary structure linear chains,
2 - 2-dimensional sheets of parallel protein threads bound laterally,
plane or pleated
3 - 3-dimensionally folded proteins, globular proteins
4 - gathered globular chains with centres of coenzymes as expressions
for the 4th
d-degree equivalent with forces. (Often
2 or 4 similar proteins together.)
5 - The cilia or centromere structure perhaps interpretable as the
5th level ?
The centromere structure (and centrioles, cilia) with central and
anticenter filaments (as 0- and 00-poles, in number 9 + 1 or 9 +
2) is probably the way the 5th d-degree is realized in a visually
3-dimensional world: The d-degree of motion, 0/00, is also equivalent
with 5', the 5th dimension degree transformed into pure motions.
Cilia represent on the macro level of cells the motional organs.
Protein fibres as collagen have also 5 levels of structure:
1. Amino acid chain - the collagen molecule.
2. Protofibril of 3 such chains spiralled.
3. Collagen fibrils with protofibrils stored both after and besides
one another.
4. Packets of these fibrils.
5. The collagen thread.
(Perhaps noteworthy too is the information that the mean molecular
weight of proteins are about 104 - 105 u à
la 4-5 ten-power steps.)
Chromosome as "lumosomes":
Could the DNA-spiral be perceived as a kind of "built-in light
wave", chromosomes as "lumosomes" on this much higher
level of complexity? Structured through the help of electromagnetic
(EM) waves?
The DNA-helix seems to have some fundamental properties
common with light beams. Its self-sustaining ability, its
self-preservation capacity, its self-supporting process, its "endogamy",
is of the same nature. As a light wave may be interpreted is self-supporting
through "breathing of vacant space" (see file about EM-waves),
levelling the negative energy of vacant space up to zero, in the
same sense the complementary principle may be seen working when
DNA-strands gets copied from individual complementary nucleotides
in the environment as anticentre, the 00-pole.
[A corresponding breathing of "emptiness"
could eventually be found in the atomic nucleus as a kind of proton
→ anti-proton-relation (an old
hypothesis among physicists). H-bridges of DNA as corresponding
to a p-/ anti-p-relation on this higher level?]
What should in that case correspond to the E- and M-factors in
the EM-waves?
On the most fundamental level perhaps the basic
NHx-groups, inwards, ~ M, versus the acidic OHx- or =O-groups outwards,
~ E ?
On a secondary level perhaps the chains of amino
acids as "radial" (~ E) versus codon bases as "circular"
(~M) ?
On a third level the polarity between U-C-bases
and A-G-bases ?
Is there any way to connect the complementarity between purine and
pyrimidine bases with the relation electric versus magnetic fields?
Codons G and A in 2nd position code mostly for
polar amino acids, U and C in 2nd position for non-polar amino acids,
hydrophobic ones, probably connected with magnetic fields, closed
structures. (?) The question is left open, but the DNA-spiral seems
more than just a metaphor for a ray of light.
It's said that a magnetic monopole (if any) should have the mass
137 times that of the electron. Mean value for an unbound amino
acid is 136,5 u. Mean value for pairs of amino acids = 273 = the
mass of charged π-mesons in electron
units exchanged in the nuclear force.
Perhaps the 4 codon bases should be apprehended similar to complementary
colours?
Three elementary colours (A+U+G) plus 1 (C) for
the formation of two complementary pairs?
Spectral lines of H-atoms:
In the "Lyman-, Balmer-, Paschen-Brackett-series" for
spectral lines of hydrogen, quotients between the spectral lines
happen to give numbers for the mass of codon bases:
The Balmer series: m = 2, n = 3, 4, 5 within the visual area:
m = 2, n = 5, 4, 3 givesλ = 4341,
4863, 6565 Å:
R(1/4 - 1/25) R(1/4
- 1/16) R(1/4 - 1/9)
→wavelength
a
b
c
a/b x 102 = 112 = U-base
b/c x 102 = 135 = A-base
a/c x 102 = 151,2. ~ G-base (151)
And what about the C-base?
The lowest spectral line of oxygen is 4368 Å. Quotient to
the the middle spectral line here for H 4863 Å = 111,3. x
10-2. (?)
[Quotients between spectral lines could in some sense be apprehended
as related to phase-waves, said to carry no information ! ?]
As mentioned before: Behind all aspects here lies the thought or
hypothesis that DNA or RNA and the coded amino acids on some deep
biochemical level made up a unit (a whole which could be called
DAP, P for proteins). An entity perhaps belonging totally to an
underground of mathematics? A whole which then got polarized through
different phases and angle steps etceteras and developed to all
processes of the protein synthesis between primary poles.
There is DNA with a protein matrix, there is rRNA
consisting of both proteins and nucleic acids and there is tRNA
carrying amino acids transported by the A-base as AMP. And the bases
are mainly constructed by and of amino acids.
To DNA-RNA-bases -
synthesis and complementarity
To Numbers of Codon
bases and some relations to numbers of amino acids
|










 tRNA 2←1
tRNA 2←1