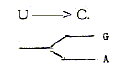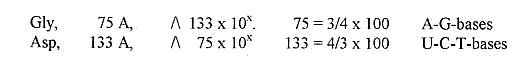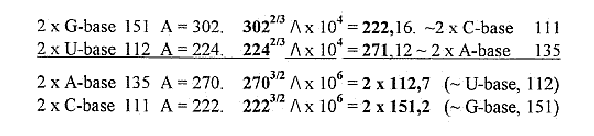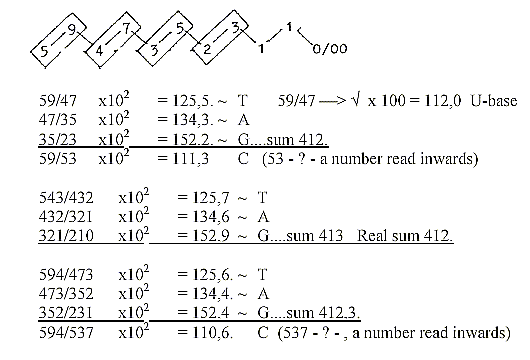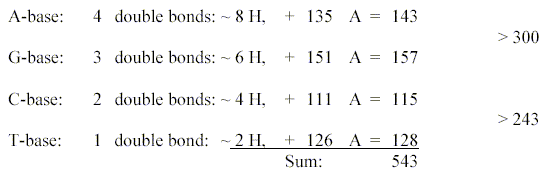|
Synthesis,
complementary traits, first annotations
about mass.
The
DNA-RNA-bases G and A versus C-U-T
do indeed represent complementary
"poles", with several
polarities in origin and their synthesis.
The
6-5-member-rings of G- an A-bases
originate from Inosinic acid,
the 6-member-rings of U-C-T-bases
from Orotate. In original
papers G and A are called "00"-bases,
U-C-T "0"-bases. Yet the
G- and A-bases in their construction
have the character of 0-poles of
the dimension model here, U-C-T
the character of 00-poles (anticentre
poles). a) G- and A-bases
are constructed from a centre outwards,
the U-C-T-bases inwards:The centre
in G- and A-bases is made up of
the smallest amino acid Gly, whose
branches outwards are filled with
small molecules from the surrounding
anticentre.
We
have a "c - ac"-relation. The U-C-T-bases on the contrary
are created through the meeting
from outwards of two bigger molecules,
the amino acid Asp and carbamylphosphate,
binding to form the 6-atom rings.
Principle of synthesis
for the opposite codon bases:
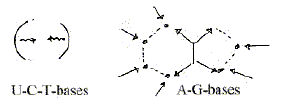
b) Further, G and A then
are formed along a branched way
from the resulting Inosinic acid,
while U, C and T are differentiated
along a "linear" path
of changes.
 c)
Also the relation to the P-ribose
groups of the nucleotides are opposite:
construction of A- and G-bases occurs
on the P-ribose group which comes
first,
U-C-T bases are constructed
separately with the P-ribose group
added afterwards. d) Gly and
Asp, the amino acids making up parts
of the polar types of bases have
mass numbers which are inversions
to one another: 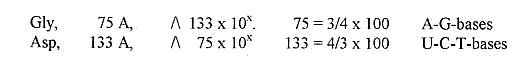
(Cf.
sum of 133 and 75 = 208, numbers
reappearing in the Exponent
series.) e)
Mass number sums of "0"-
and "00"-bases are inversions:
G + A = 286 =
151 + 135 A (with +1 for bonds to
ribose)
U+C+T = 349 = 112 +
111 + 126 A
286
/\ 349. x 10x
Also, more intricate:
2 bases with exponent 2/3 give their opposites if inverted.
(Cf. the exponent 2/3 to an elementary number chain 5-4-3-2-1 related
to codon distribution to amino acids in the genetic code, see files
The genetic code.)
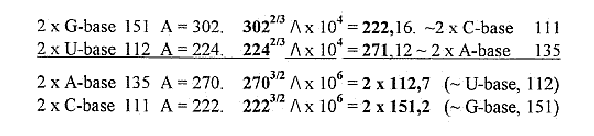
The branched paths of synthesis
for G- and A-bases:
Here
we have something like a secondary
polarity: A-base gets its
free N-group from the amino acid
Asp,
the
N-group in the B-chain of this.
(There is of course no other.)
G-base gets its free N-group from
Gln,
the
N-group in the R-chain of this.
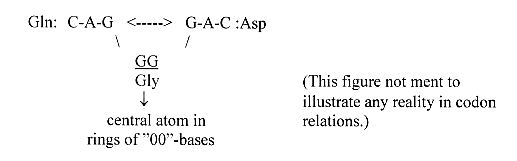
We can note too:
Asp,
70 Z ——> A-base 70
Z
Gln, 78
Z ——> G-base 78 Z Codons
as a mirror relation: Asp
G-A-C <——> C-A-G
= Gln
(but G-A-U <——>
U-A-G = one of the Stop codons.)
Complementary
forms of bases versus amino acids
(ams):
There is a relation
in their forms like centres-anticentres,
(c—ac), as 0-00 in the model
- or radial versus circular as geometrical
poles of dimension degree (d-degree)
3 in our model:
- Amino acids
as tetrahedrons have central C-atoms
and direction outwards in 3-4- directions,
- Bases are rings as if the central
atoms had got inverted to anticentre
positions. Formally, 2 amino
acids N-C-C-N-C-C could be restructured
to the 6-rings of the bases (if
chemically possible is another question):
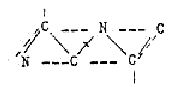
Actually,
the contribution of the amino acid
Gly to A- and G-bases corresponds
to half a 6-ring (1/3 of the C-N-atoms
in the 5-6-ring), and Asp with C-C-C-N,
2/3 of the 6-rings of U and C, T:
7 out of 15 C + N in purine and
pyrimidine rings.
(Also
Glu and Asp contribute with singular
atom groups (NHx) to the Inosine,
the parent to A- and G-bases.) Polarization
in end-(R)-groups of the bases:
In the G-C-pair the bases have both
O and N, keto-oxygen and nitrogen
groups,
while the opposition
O-N is polarized to separate bases
in the A-U-pair (A-base with only
a NHx-group, U (and T) with only
2 oxygen groups.
This
fact points towards the A-U-pair
as expression for a secondary step
or level from the viewpoint of our
dimension model. Many other facts
do the same in the arithmetical
analysis of codons and amino acids
(see those files). (That the opposite
is true about the order of synthesis,
representing the opposite inward
direction, is natural.) The
base pairs may be ordered along
3 kinds of polarities: a.
GA <——> UC type
of bases
b. GC <——>
UA complementary pairing
c.
GU <——> AC defined
as keto-bases versus amino-bases.
The GC <—>
UA- polarity is here suggested as
the first one, an expression for
opposite, complementary directions,
i.e. d-degree 4. The GA <—>
UC-polarity could be the second
one. Compare features of "radial"
versus "circular" in the
forms of synthesis as poles of d-degree
3. Also a certain polarity heavier
— lighter in the property Mass.
The GU <—> CA
polarity, founded in the O <—>
N-polarity, refers to a hydrophilic-hydrophobic
aspect, which concerns Charge as
a property. Charge in this model
assumed as a property defined in
d-degree 2.
Illustrated as
coordinate axes:
 3/2/relations
among bases as a polarization of
5: 
Free end-groups of the 4 RNA-bases
as illustrated in end-groups of
amino acids Gln and Asn:
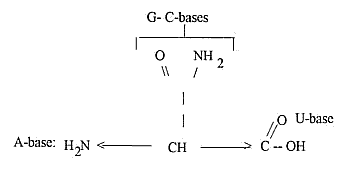
About
mass numbers (A) and atoms in the
single bases: Intervals
in mass between A-T-G-bases:
Intervals are squares illustrated
by the Pythagorean' triangle.
G 151 u, A 135 u, T = 126 u. (Including
+1 for bond to (deoxi-)ribose: 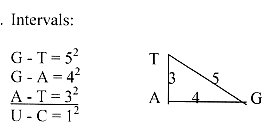
Approximate mass numbers of T-A-G-bases
from quotients in the dimension
chain with superposed odd-figure-chain:
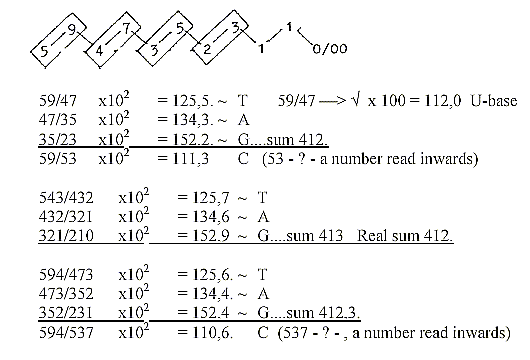
Sum of the 4 DNA-bases with +2H
for the double bonds added = 543:
543: the first triplet in the
elementary number chain:
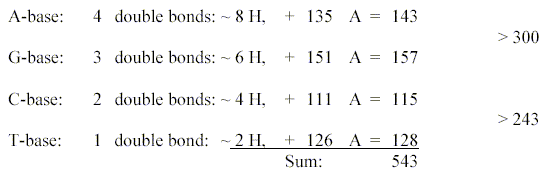
[Mass of side-chains of G+C-coded
ams (R) = 544. The same number division
appears there between Z- and N-numbers:
300 Z, 244 N (-/+1 in 2nd base order)]
A mass relation between base
types as connected with sum of ams:
Pyrimidine ring without H: 4
C + 2 N = 76 A
Purine ring without
H: 5 C + 4 N = 116 A
76/116 = 2 x 3275,9. ≈
3276. x 10-4. 3276 = sum of 20 + 4 double-coded ams.
A - Z -
L/Lp -
p - numbers as different levels
in the bases:
The starting
point here is the thought that activation
("excitation" ?) of an
atom or molecule may imply a stepwise
approach towards more superficial
levels as suppression of deeper
ones and therefore different number
of levels in the electron shells
are engaged in different stages
of processes.
Here
we assume that in first steps the
orbitals of the K-shell are suppressed
first, designated L-level, in next
step the s-orbitals of the
L-shell, designated p-level.
It doesn't include H-atoms of course.
Addition of different stages
could then be regarded as an operation
for getting the time aspect inherent
(only virtual). 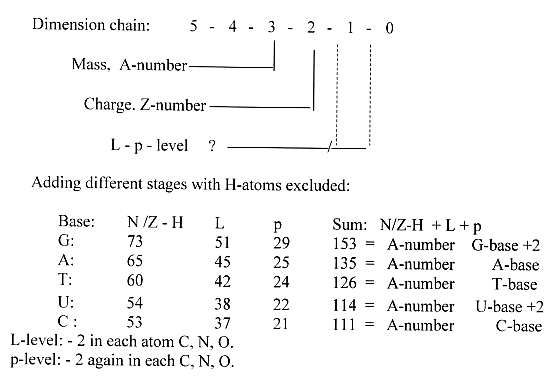
Here it seems as the deeper level
of atomic Mass is "disintegrated"
in different stages of more superficial
levels - as Charge and the electron
shells - in a way that agrees with
the general principle in the dimension
model of higher d-degrees transformed
to lower ones. There is a similar,
rather remarkable pattern among
the coded amino acids, see The
genetic code.
Number of atoms in
2 x 24 codon bases, 1st and 2nd
position:
Cf. table
of 20 + 4 double-coded amino acids.
- Equal number divisions +/-1 in
2 dimensions, horizontally an vertically.
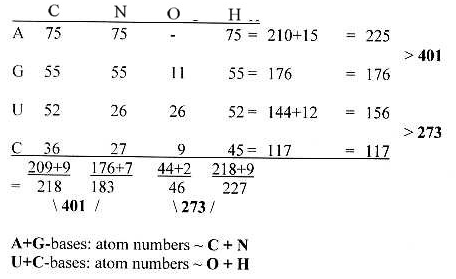
Total
sum of atoms = 674 = 2/3 x the sum
of the
exponent series
1011:
With 3rd base in codons
included, assuming equal distribution,
6 of each base, 336 atoms are included:
6 G = 6 x
16 = 96,
6
A = 6 x 15 = 90,
6
U = 6 x 12 = 72,
6
C = 6 x 13 = 78
Total sum of atoms should then become
~ the sum of the exponent series
1011. 52/3
—
42/3 —
32/3—22/3 —12/3
—0
x
102 = Sum 1011, 3 x 337.
Numbers
abbreviated to integers in the exponent
series: 292
+ 252 + 208 + 159 + 100
544
467
Atoms
then in the bases paired:
A+U-bases:
544 atoms -1 = 292 + 252
-1
G+C-bases: 467 atoms
= 208 + 159 + 100
Including
stop codons, UA-A/G and UGA, first
two bases = + 2 U, 1 A, 1 G:
The same equivalence vertically
and horizontally appears +/-1:
A+G-bases:
432 ~ C + N = 433.
U+C-bases:
297 ~ O + H = 296. 23
amino acids, without Ileu2 ( same
1st and 2nd base in the codons): 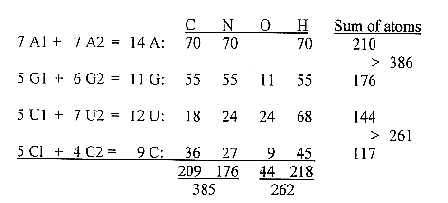
a.
A+G-bases: atom number ~ C+N atoms
(A ~ C, G ~ N)
U+C-bases:
atom number ~ O+H atoms. b.
Number 385, which guide one 12-group
among amino acids, is equally divided
here
in
the numbers 209 - 176 (among the
Cross- and Form-coded ams +/-1).
Numbers 262-261 is the
mass number of a DNA pair G + C
and A + T respectively.
Note
perhaps the inverse relation between
numbers around 260 and 385.
There is also the relation in number-base
systems (nb-x):
261
in nb-10 = 405 ~ 385 rewritten in
nb-8.
(From files The genetic
code.) The schemes above
seem connected with the function
the bases have as coenzymes in relation
to the different classes of substances.
Roughly:
U-base
(UTP) with carbohydrates (dominating
atom O),
C-base
(CTP)- with lipids (characterizing
atom may be said to be H)
G-base
(GTP) with proteins (typical atom
N).
A-base, less specific,
most connected with energy storing
and transportation.
Sum
of products in the dimension chain
with superposed odd-figure level
gives mass numbers of bases and
amino acids:
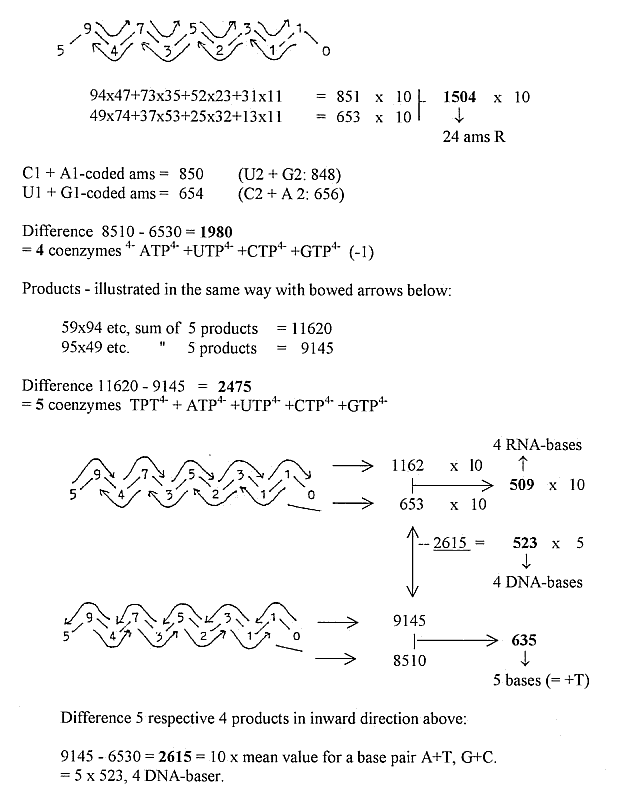
A-base and nucleotides
- some number readings in the elementary
chain:
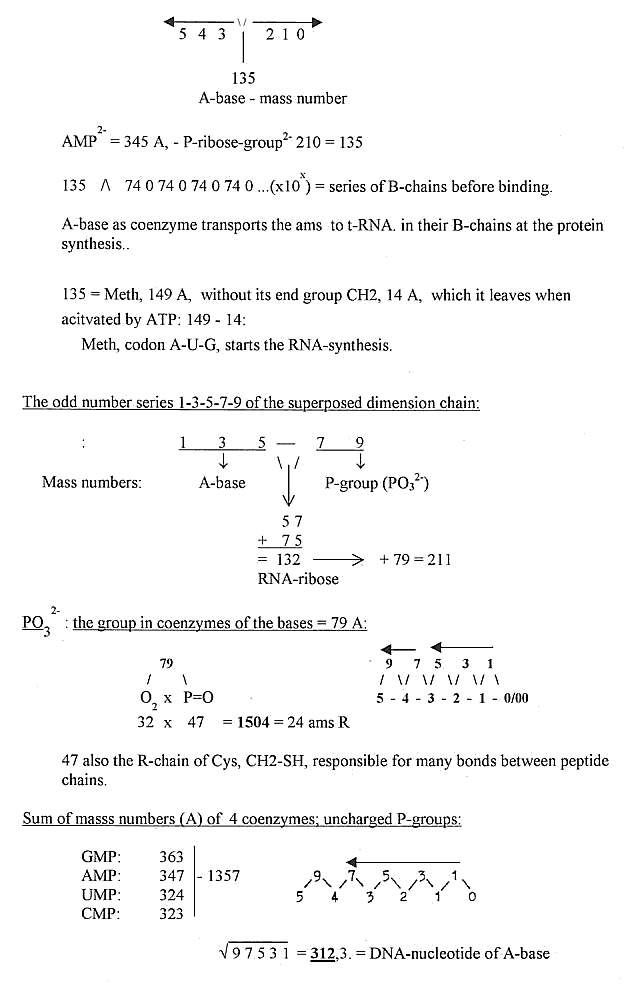
* To
Numbers
- more on mass numbers of bases
and mass relations to amino acids.
|