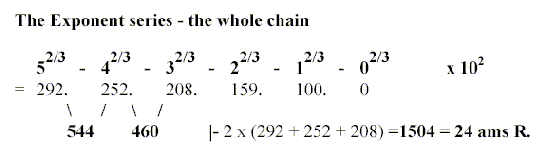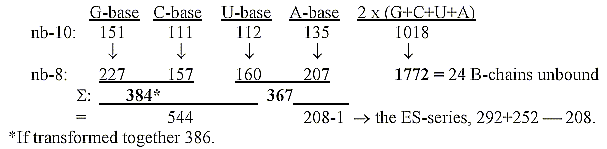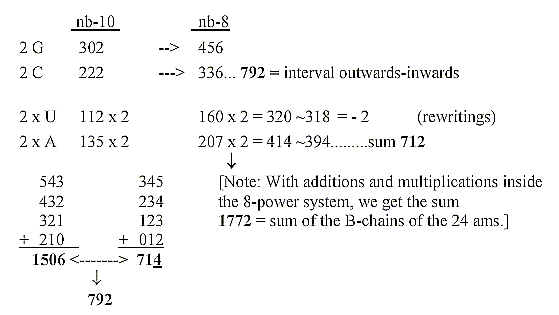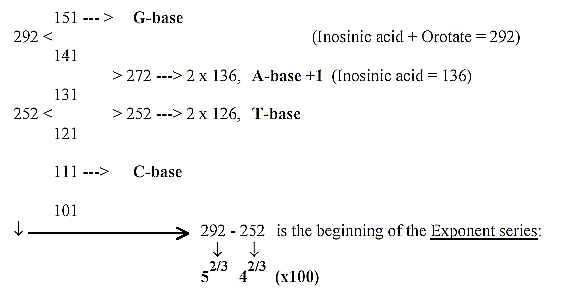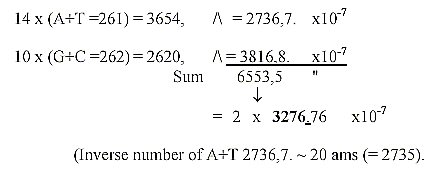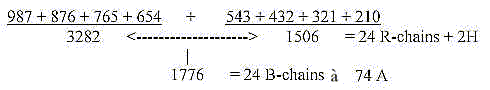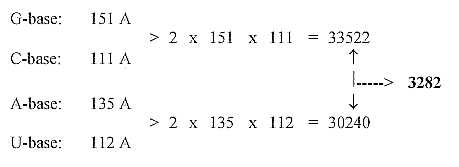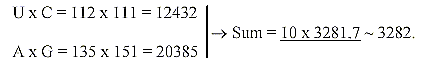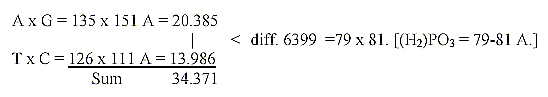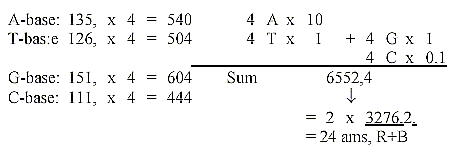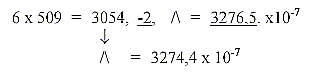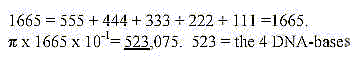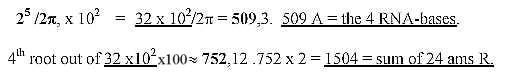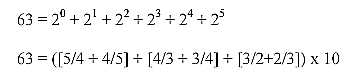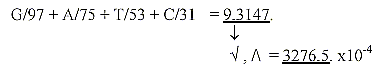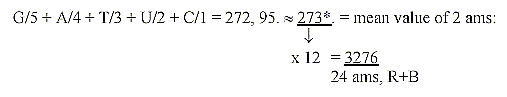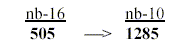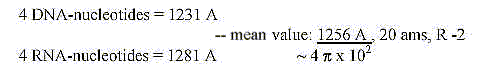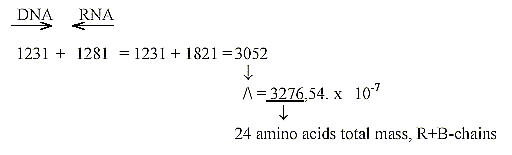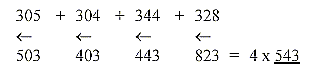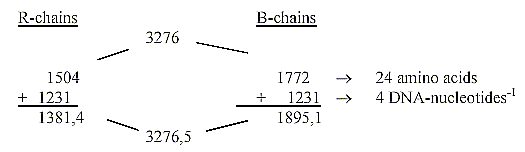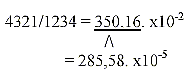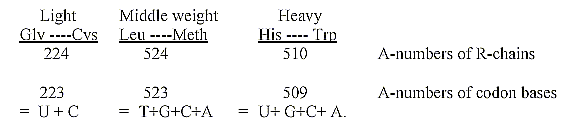|
Used abbreviations:
Ams = amino acids.
R-chains = the different side chains of ams.
B-chains = the similar part, which bind through condensation.
- Here it's calculated with 24 ams, the four double coded included,
where nothing else is mentioned. See table over ams here.
/\ , sign indicating inversion.
nb-x = number-base system, e.g. nb-10, nb-8 etc.
1. Two sets of the 4 bases give the mass sum of
coded amino acids in nb-8:
a. The elementary number chain 5 - 4- 3- 2- 1- 0 with
exponent 2/3 x102 called the ES-series:
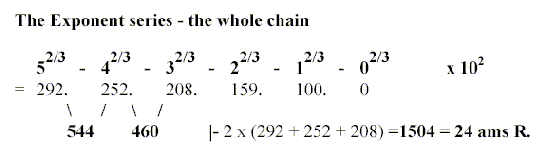
544 - 159 =385, 208 + 159 =367.
*Numbers 292-252...etc. below referred to as "5", "4"
etc. elementary figures within quotation marks.
Transformations of base numbers to nb-8:
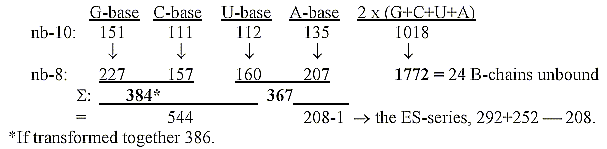
2. Division of the sum 1504 in 714 - 792-2:
Mathematical operations performed in nb-10.
Sum of 24 ams, R-chains = 1504 divided in numbers
792-712,
related to the sums of triplets of a dimension chain through rewritings:
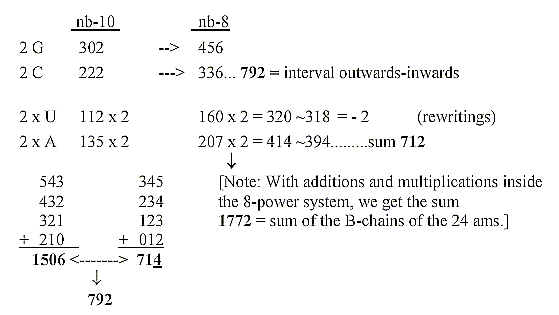
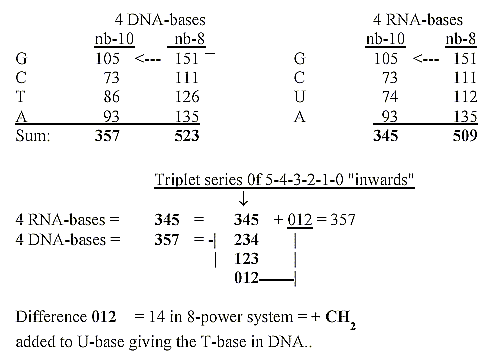
3. Mass numbers of bases read as in nb-8, separately translated
to
nb-10 - and the triplets of a dimension chain:
4. Mass numbers (A) of the bases read as 6-power numbers:
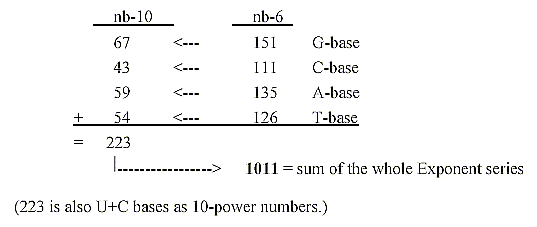
5. The bases bound with exponent 2/3 give numbers of the ES-series:

6. Middle figure in the A-number of bases as a dimension chain
between "poles" 1--1 as in the last d-degree of motions
in the model:
Numbers 292 - 252 from the ES-series:
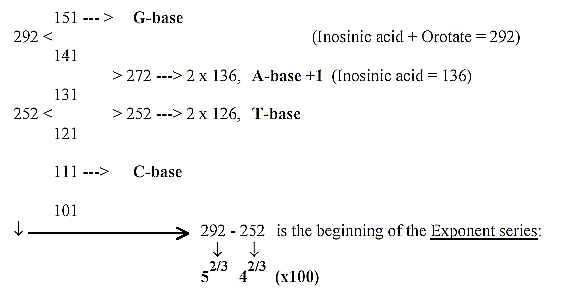
With 2 times 131 + 121 + 111 = + 363 = 655 + 363 = 1018 = 2 x 509,
the sum of 4 RNA-bases. All these numbers transformed to nb-8 give
the sum of the 24 ams divided 973 - 531 (as triplets of the odd-figure
chain 9-7-5 (-2) —531.
Products between bases
7. Products between bases and (approximate) sums
of amino acids:
Number of bases in 1st and 2nd position times A-numbers of the pairs,
inverted:
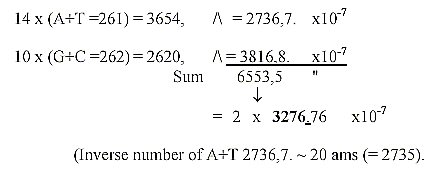
8. Products of bases and number 3282 - 3282 as an approximation?
The triplet series 543-432-321-210 expanded gives approximately
R+ B-chains of the 24 amino acids:
Sum of 24 ams, unbound, R+B-chains, = 3276: There
is a loss of one H in B-chains of 4 ams, Arg 1, 2, Lys, Pro. when
Arg and Lys have charged N-groups in the R-chains.
Could there eventually be a stage where there is +2H in some R-chain,
giving the sum 1506? Cf. the triplet series of the dimension chain:
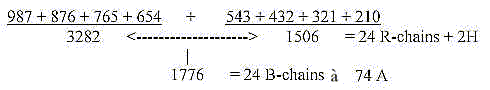
1776 = B-chains before reduction of 4 H in B-chains
of Arg1,2, Lys, Pro,
Difference between base products of base pairs:
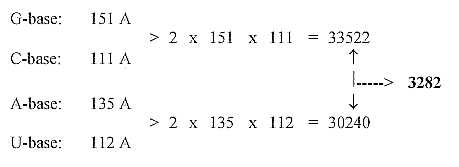
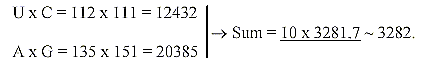
9. Products of bases DNA divided with 4 π:
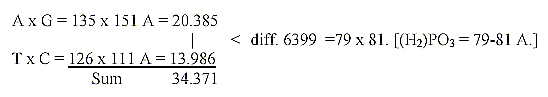
Difference 6399 / 4 π
= 509,2. 509 = A+U+G+C, the 4 RNA-bases
Sum 34371 / 4 π = 2735,15. 2735 = 20
ams, R+B
Various other kinds of operations.
10. Four times the base number with displacements
in the 10-power positions:
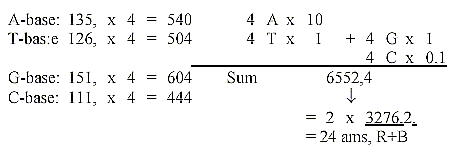
11. 24 RNA-bases, as if equal use in the codons in one position:
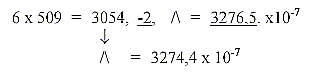
12. Natural logarithm e:
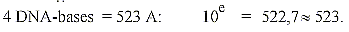
13. Dimension chain numbers as n x 111-numbers,
times π:
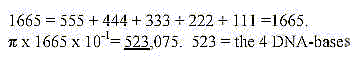
14. Number 32 and connection with the π-number?
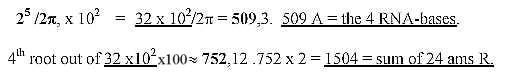
[Quotient 105 / 25 = 3125
→ log 3125 = 3,49. Inverted ( /\)
= 286,1. x 10x.
Cf. the hypothesis in the model that 10 could be the log-base
as sum of poles in d-degree 4 and 2 the log-base in polarizing
direction as sum of poles in d-degree 0/00.]
15. How to show that sum of the 4 bases ≈
31-32:
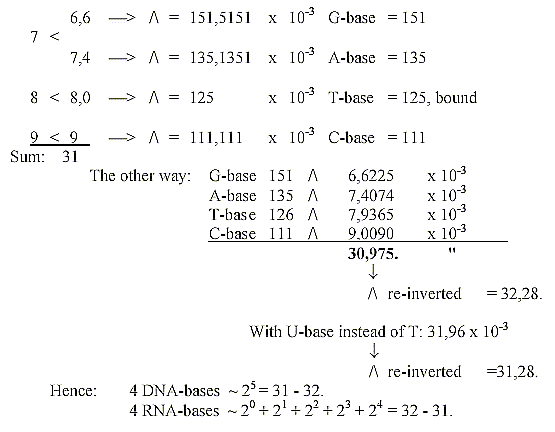
16. Number 63:
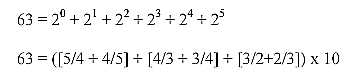
- Sum of 5 bases bound = 630 = 1/2 x 1260,
- (6,3 x 4)2= 635,04. 635 the sum of 5 bases.
- Mean value of a base = 126 A = A-number of the T-base unbound.
- Mean value for a side chain of ams ≈
½ x 126 = 63 (62,75),
counted on 20 + 4 double-coded ams.
- 1260 - 2 = A-number for side chains (R) of 20 ams.
- 63 = the difference A+G <—>
U+C = 286 - 223.
- 63 = PO2 -group, connecting the nucleosides
in DNA/RNA.
17. Bases divided with number steps,
read in the odd-figure chain 9-7-5-3-1:
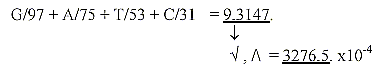
18. The 5 bases according to their mass: G-A-T-U-C divided with
the elementary number chain 5-4-3-2-1:
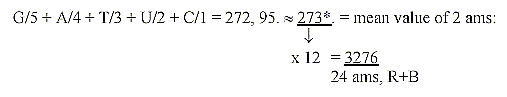
*A+G = 286, T+U+C = 349: 286 /349≈0,819.
819 = 273 x 3 = 1/4 x 3276, sum of 24 ams R+B
19. A more odd operation:
First figure in the A-number of the bases regarded as detached
from the last figure:

Nucleotides — and
mirrored numbers
20. Triplet of nucleotides in RNA with mean value 320 - 321
= ¨960 - 961:
Cf. sum of triplets in the elementary chain: 432+321+210 = 963.
960 = A+U-coded ams R.
21. The 4 bases bound = 505 give the sum of their nucleotides
through nb-transformation:
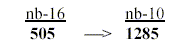
22. Nucleotides with P-groups charged -1:
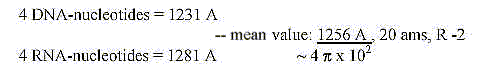
With uncharged nucleotides 1260 A: 20 ams R = 1258
≈ the middle value.
23. Mirrored number relations?
a. Mass sum of RNA-nucleotides read backwards as in
a mirror relation to DNA:
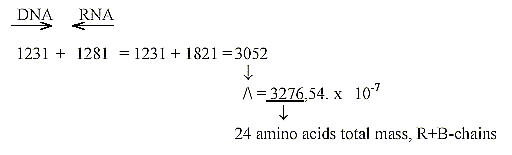
b. Mirrored numbers for separate nucleotides RNA:
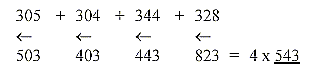
543, the first triplet number in the elementary dimension
chain.
543 also the sum of the 4 bases when +2H for each double-bond in
the rings are included but this concerns the 4 DNA-bases.
24. Amino acid sums, R and B chains added with
displaced DNA-nucleotides:
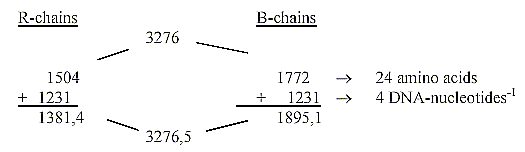
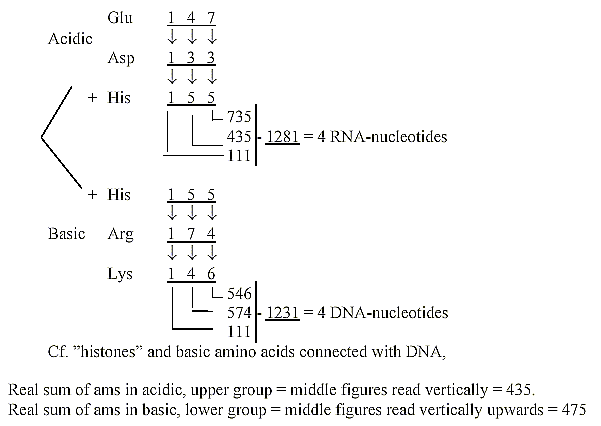
25. The four DNA- and RNA-nucleotides 1231-1281:
The sums derived from an angled reading of acidic and basic amino
acids
(R+B-numbers), His included in both groups:
Miscellaneous other operations
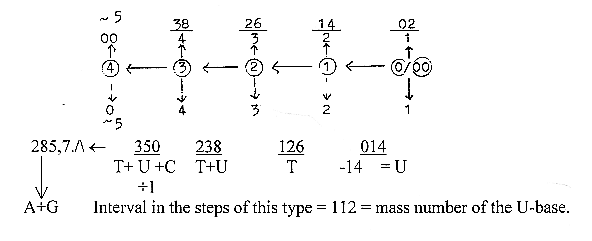
26. The sums of 2 purine and 3 pyrimidine bases,
286 and 349;
a. Numbers for dimension steps "inwards" with sum of
poles of next higher degree added, for instance step 4 ← 3
= number 3-4, + poles 0 and 00 worth 5 = + 10, read as 350:
b.
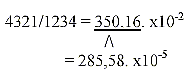
27. Division of 20 ams in accordance to their weight in three
groups gives sums which -1 corresponds to grouped bases.
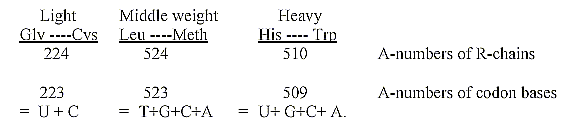
28. Numbers from 2-figure-reading and additions
in the "2-figure-chain":
Example: 59 + 94 + 47 + 73 + 35 = 308 etc.
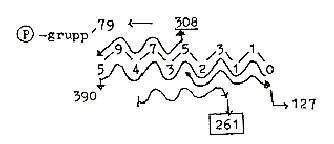
308: Mean value of a DNA-nucleotide = 307,75. (charged)
261: Base pair A+T.
127: Mean value of 5 bases unbound = 635/5- = 127
29. Mass number of the G-base 151 from inverted
triplets:
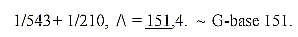
30. Mass number of C-base 111:
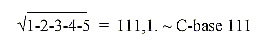
31. Rolled numbers:
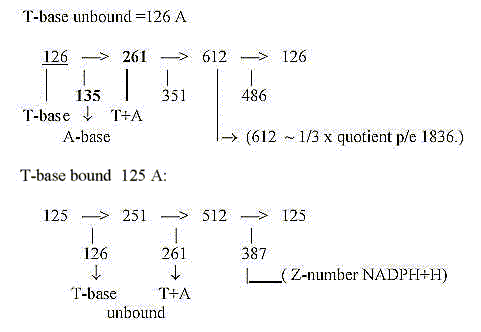
*
|