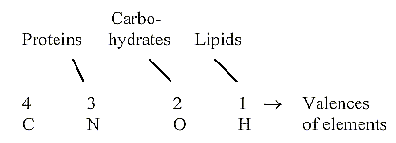|
Some tested aspects from the viewpoint of the
dimension model.
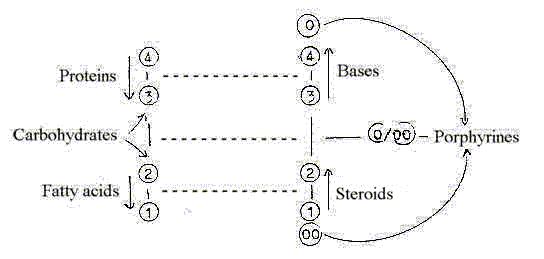
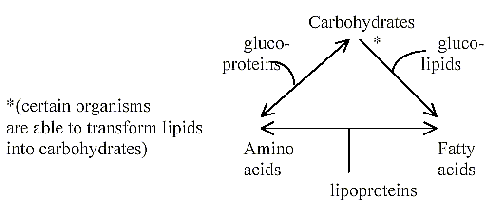
1. How many main classes of substances to count on? 3-6 ?
- Carbohydrates — Porphyrines
- Proteins — Nucleic acids, bases
of the genetic code
- Lipids — Steroids (isoprenes)
We can try regarding the left and right classes as complementry
pairs:
The classes as "poles" in a 3-dimensional co-oodinate
system?
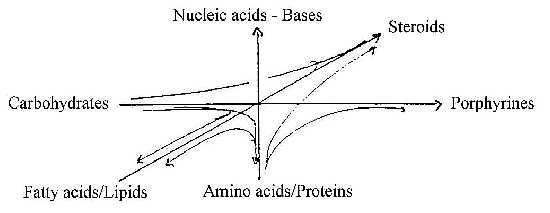
Connection between the classes in pairs:
Carbohydrates - via keto acids →to
Succinyl~(CoA) → to Porphyrines,
Porphyrines → chlorophyll→
photosynthesis → synthesis of carbohydrates...
Proteins, amino acids - via Asp and Gly →
to synthesis of pyrimidines and purines, the nucleic acids, U-C-T
and A-G. (Asp ~ ½ of U-C-bases. Gly starting centre
in A-G-bases) → to protein synthesis...
Lipids, fatty acids one branch from Acetyl~ , the other branch
to isoprenes, steroids, carotenoids, uinones.
The polarity between open ("radial") chains and closed
("circular") rings may be observed, as one feature of
complementary poles in the dimension model:
(Condition: carbohydrates divided and transformed
to keto acids.)
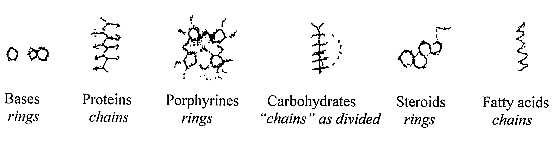
Interpretation of the 3 "coordinate axes" in a dimension
chain as 3 levels ?
Level III
Lipids Steroids
Level II Amino
acids Bases (nucleic acids)
Level I Carbohydrates
Porphyrines
Here we see the closing to ring forms as following from steps in
lower d-degrees. (The order here of levels I-II-II corresponds
to characteristic atom kinds O - N - C and their "A-Z-numbers".)
From other aspects, the order of level I and II should be
the opposite.
2. Classifications from aspects on elementary molecules:
2.1. D-degree of main form character:
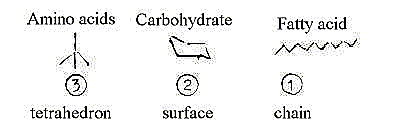
Inward direction: fatty acids - hydrophobic
Outward direction: carbohydrates - hydrophilic
Secondarily polarized: amino acids of both kinds: hydrophilic/hydrophobic
2.2 Characterizing elements - valences:
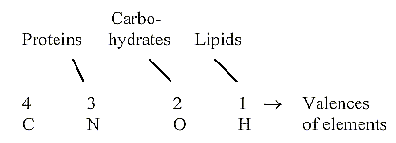
Forms of the molecular parts: 3-2-1-dimensional;
Roles when bound together in macromolecules:
4-3-2-dimensional.
Thus, the synthesis of the small molecular parts implies
an increase of the dimension degree with 1 step :

(This aspect is of course a simplification. Amino acids for instance make up also structural part of d-degree 3, the radial 3b-pole in the model here. And lipids may be seen as characterized by the C-atom and have a more high degree role than usually are attributed to them?...)
2.3 Genes - proteins - carbohydrates - lipids in the cell
as a "fruit":
Growing and increasingly circular potentials towards
lower d-degrees as one hypothetical aspect on a dimension chain:
Core - pulp - peel: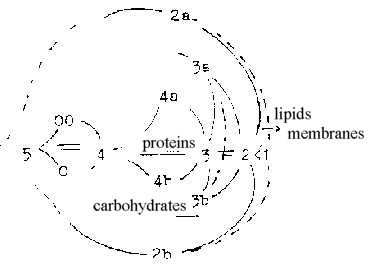
2.4 Ring forms as through inward direction in d-degree steps,
chains as outward direction in the pair of classes respectively?
4-3- and 2-1-steps: chains in outward direction, ring-forms
in inward direction?
3-2 step: as double directed: carbohydrates.
0/00: outer poles meeting as 5', porphyrines.
2.5 The treatment of water:
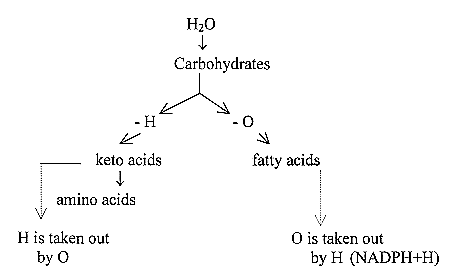
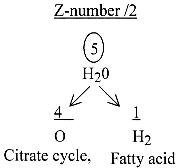
Inside/outside mitochondria:
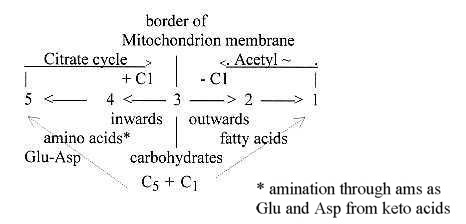
2.6. Synthesis in number of C-atoms as an aspect:
O: Carbohydrates C5 + C1 →
C3 + C3 (Pentosephosphate cycle: 3 C5 →5
C3)
N: Amino acids C4 + C2
→ - C1 →
C5 Glu (from citrate cycle); also ams from C3 molecules in glycolysis...
C: Fatty acids C3 -
C1 + C2 x n.
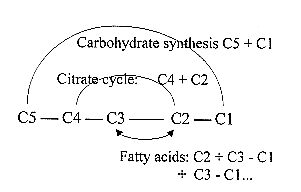
Fatty acids: inverted direction in middle-step 3←
2.
- Pentosephosphate cycle 3C5→
5C3: compare sum 15 of the elementary number series.
Middle level C4 + C2 → Citrate
cycle - keto acids
↓
Succinyl~
+ Gly = C4 + C2,
↓ - C1
Porphyrines
→ Chlorophyll
A note about porphyrines and their "side-chains"
or "rope ends" at the rings:
- Rings: C5 x 4
Chain ends 4
x (C3 + C2) before turning
becomes
1 x (C3 + C3), 2 x (C3 + C2), concerns Uroporphyrinogen
III),
becomes 1
x (C3 + C3), 2 x (C2 + Cl), 1 x (Cl+CI),
this
a principal in Haeme and Chlorophyll.
Reduction/cut/diminution of chain ends as a kind of step displacements
?
Cytochrome a: addition of an isoprene chain (isopentenyl~).
2.7 More complex molecules, the combinations:
3. A dimension chain with elementary physical concepts
as suggested in files about Physics, connected tentatively
with classes of substances:
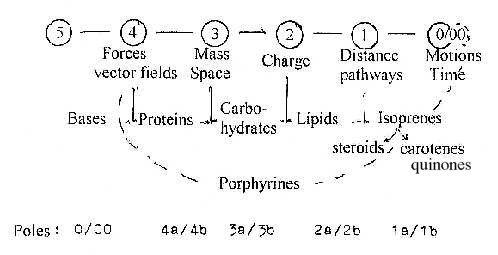
The figure refer to a mix of aspects: connections
agree approximately with forms and functions in the cell. (Lipids
as responsible for potentials +/- over cell membranes.)
However, connections don't agree with the view
of polarization steps outwards in the chain: Bases don't "polarize"
into proteins even if they guide the differentiation of amino acids.
And proteins assuredly don't polarize into carbohydrates but individual
amino acids "break down" to different stations in glycolysis
and citrate cycle and most steps in these are double-directed. Hence,
proteins may this way be transformed to carbohydrates. With the
aspect of synthesis, this appears double-directed inwards and outwards
respectively.
Most obvious example of a polarization giving complementary
"poles" as partial structures in accordance with the
dimension model is naturally the division of carbohydrates (hexose Glucose
→ Fructose) leading to amino
acids (proteins) and fatty acids (lipids) respectively: Two forms
making up radial versus circular parts in the cell as poles 3b versus
3a.
(We may regard the radial 3b-pole as derived from
the 0-pole in a haploid chain, the "circular" 3a-pole
as derived from the 00-pole:
3a
<— <— <—- 00
|
0 —> —> —> 3b )
To get this polarization - regarding direction of
synthesis - in some correspondence with the figure above,
a version of the loop model seems needed, the three polarization
steps 5→ 3/2, 5 →4/1,
5 → 0/00:
.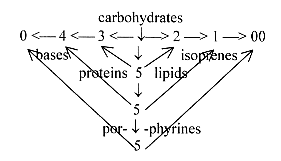
[Cf. in the figure vertically 5-5-5 and Pentosephosphate cycle 3
x C5 → 5 x C3.]
Proteins: step 4-3, poles 4a/4b:
- Amino acids which in synthesis ( ~ inward direction) give parts
of the codon bases.
- Enzymes as forces, polarizing / binding as 4a/4b-poles,
(- L/D-forms of amino acids (example of secondary double-direction),
where one direction has been selected as dominating in higher species.)
Carbohydrates; step 3-2, poles 3a/3b;
- Number of C in pentoses 3 + 2, in glucose 3+1+2: The 6th C-atom
gets bound in the middle of the C-chain. Hexoses divided C3/C3 as
poles 3a/3b.
- 3a as the part transformed to glycerine, building "circular"
backbone form of lipids,
3b developing to Pyruvate and Acetyl~ C2, starting synthesis of
fatty acids.
- Also another polarity on macro-scale: 3a as cellulose type, with
role as "spherical" coating, 3b amylos or glycogen as
stored substrate in the cells, a complementarity similar to the
type space / mass in the model.
Lipids, 2:
- Synthesis from the parts of carbohydrates, the poles 3b/3a.
- Number of C-atoms in syntheses: 2 + (3 -1), x n, Plus C3 for the
backbone to triglycerides."Linear" chains
coupled to glycerine into 2-dimensional forms.
- Roles in membranes as demarcation, "shell", surfaces,
d-degree 2.
- Charge (assumed as property defined as such at d-degree
2) across the membrane: negative inside, positive outside, inversely
to the distribution of charge of atoms.(! Life as antimatter on
a higher level! Cf. life as processes, the dimension chain in opposite
directions to structure degrees.)
- The 2/1-division of triglycerides in directions from binding backbone
(glycerine) to P-lipids and more complex .molecules as glycolipides.
Ring structures:
Isoprenes
→ (squalene) →
steroids → carotenoids →
quinones: 2 → 1 →
(0/00):
This class with the derivatives of isoprenes illustrate in many
ways the d-degree steps 2 →1
→ (0/00, the porphyrines).
- Synthesis of an isoprene as of fatty acids from Acetyl~,
C2 but from 2 x C2 plus a third branch C2:(as a step 2 →2a/2b)
C2
+ C2
|
+
C2
- Lipids versus isoprenoids: two different ways to form 2-dimensional
macromolecules: in lipids: 2 crossing coordinate axes, in steroids:
wavy forms of chains closing to rings in opposite directions. This
opposite use of C2-molecules seems to reflect a polarity of the
kind radial versus circular in the synthesis itself.
- The wavy form of squalene as oscillation forms of max/min, convex/concave:
equivalent with (~) poles 2a/2b, closed to steroids: flat planes
(2).
- Steroids among other things parts of membranes, hence closely
related lipids.
- Functions: among other things as hormones, the chemical signal
system. Connected with light, electromagnetic waves, as in D-vitamin.
In these functions also representing less of structure building,
more of motions as in the polarity particles/waves in physics.
- Further carotenoids, xanthophylls
connected with organelles for photolysis and a derivative as visual
purple (rhodopsin) for light absorption in eyes. Long "linear"
chains with ring-closed ends as in d-degree 1 with poles: 2a----1-----2b.
Note the complementary directions of ring bows at the ends.
- Quinones: 1b ----- (0/00): Only "1-dimensional"
isoprene chains, here connected to a ring from the amino acid Tyr
(side chain 107 A), as from a 4-1-loop from proteins to last step.
- Function: Ubiquinone for instance: directly part of the "pathways"
(cf. Distance), "elevator ropes", for photon energy and
H2-transportation in the respiration cycle.
Porphyrines, 0/00:
- The synthesis giving a picture of inward directed vectors, pole
4a (Gly and Succinyl~) under rotation pointing to location for the
metal ion. In some sense similar to "black holes" in macrocosm,
"catching" light energy. (Outer poles of d-degree 4 =
0 and 00, from polarized d-degree 5.)
- As chlorophylls key substances in the big loop photolysis - respiration
cycle (vegetative/animal worlds).
- In type of form c/ac-figures, centre - anticentre, 0 and 00-poles.
The polarity on the more fundamental, underlying level of chemical
elements, appears here inverted: Non metals as representing
outward directions from 0-pole form here anticenter (from 00-pole),
while metals representing inward direction from 00-pole become centres.
(Such a pole exchange has been assumed in he background model. Also
in agreement with the general principle of stepwise building-in
of the environment as anticenter. )
Codon bases in nucleic acids, the building stones for the genes:
- Responsible for integration of the "whole" (d-degree
5) , the unity of a cell.
- In the figure above proposed as representing a step 5 ←
4.
- Constructed by amino acids and some other molecules, as in synthesizing,
inward direction, associated here with ring forms.
- Are 5-4 in number.
- Represent the most typical complementarity in a pair relation
as poles of a dimension degree in our model.
- Synthesis characterized
by centre — anticentre respectively, the partial structures
out of polarized d-degree 5.
- In one sense it's the polarization of base pairs to one-way direction
which leads to the proteins, the class associated with d-degree
4-3: the separation of the double helix of DNA for copying of mRNA.
- The "haploid" role becomes a condition for the function
of bases as "forces"in their role as coenzymes. Cf. that
one-way direction is a condition for a force to be acting. Balanced,
opposite forces cancel each other.)
- The "outer poles" (or partial structures) of d-degree
4 meet in the last step, in "d-degree 0/00" (of motions)
to which the class of porphyrines has been attached here. DNA is
also present in chloroplastes with chlorophyll for photolysis as
in mitochondria with cytochromes for the respiration cycle. And
in both Gly contributes at the synthesis, the simplest amino acid.
(Gly as outward vector from 0-pole, centre, in G- and A-bases, as
anticentre (?) vector inwards in porphyrines when combined with
Succinyl~ chains).
8. Application of other aspects on a dimension chain:
a. Increasing one-way direction toward lower d-degrees.
One possible aspect is the number of bond directions of the macro-molecules:
- Proteins, folding or not, with bonds in at least 3 directions,
surely often 4 as coenzymes. - Carbohydrates with bonds in 2-3 directions
(3 for instance in glycogen with branched chains).
- Lipids with bonds in 1-2-directions (2 with glycerine).
b. Motional moments as Vibration - Rotation - Translation in
3 dimensions:
The aspect in the dimension model that d-degree
of motions increases when d-degree of structure decreases
in steps towards lower degrees have been dealt with in files about
physics, applied to the atomic level. Surely it seems silly
trying to apply this elementary hypothesis about motions, built
on 1-2-atomic molecules, on the very complex biochemical level.
Yet, here some views on the molecules from a similar aspect.
- Protein chains in the muscle fibres (actine/myosine) and
their motion in/out may be regarded as a vibration (through
pacing by hooks on the chains), an 1-dimensional motion as
assumed in d-degree 4. (Muscles the organ for motions.)
Perhaps the function of protein microtubules in
cilia, elementary "linear" organs for motions, could be
regarded as a substantiated form of linear vibration? (The motions
of kinesin and dynein as "motor proteins",
"walking" on tubules, see Wikipedia.)
The first folding of protein chains on the underlying,
elementary level could possibly be interpreted as a stepwise 1-dimensional
motion through the 3-dimensional space.
Surely however, a lot of other aspects on protein
motions could multiply the degrees of motional patterns.
- Motional pattern of carbohydrates?
Do the macromolcules of amylos, glycogen or starch rotate, a 2-dimensional
motion?!
( The spirals of amylos implies rotation in the
structure, the branching of glycogen may be regarded as "translation
in 3 dimensions", but these are examples of patterns in
the synthesis. They don't correspond to motional patterns for the
macromolecules, the aspect here.)
- With lipids associated in structure with d-degree step
2 (then 2-1 in elementary form) )the motional patterns should be
3-4-dimensional. The amoeba-like motions of membranes,
in- and out invaginations, could be identified as an expression
for such a 3-4-dimensional motion. Cf. in structure poles 4a/4b
defined as outward/inward direction.
- About steroids as 2 ←
1-dimensional in structure and often attached and integrated in
lipids, they assuredly move around 3-dimensionally in inner
space - as sex hormones for instance, but also take part in the
regulating of gene activity. Cf. the connection 4 - 1 in the loop
model which should give a connection between genes/bases or repressors
of proteins and steroids in the figure above. If this latter function
could be interpreted as an expression for a 4-dimensional
motional pattern is debatable. Originally "pumping" has
been proposed as the form of 4-dimensional motion. Maybe also a
typical vector character (with address) of a motion could be regarded
as 4-dimensional?
Long chains of isoprenes (polyprenoles) transport
carbohydratepeptides through the cell wall in bacteria.
- A "5-dimensional" motion, when
structure d-degree is zero (in d-degree 0/00) is in the model presumed
as only the "pole exchange", the "germ"
to Motions in itself. It could perhaps be identified as just the
mentioned positioning of metal ions in the centre of porphyrines?
And/or only with an internal change in charge, in electronic energy.
In other aspects the porphyrines interpreted as principally 5-dimensional
structures seem to be characterized of great immobility.
c. Higher d-degrees defined as binding forces in relation to lower
d-degrees as structures:
Is this postulated definition in the model in any sense applicable
to the classes of substances? "Binding" (sign <) -
in which sense? And which molecule binds which in an addition?
Generally it may be said that bases in the form
of coenzymes with their P-groups (valence 5) binds many other substances
and that proteins as enzymes act as both binding and polarizing
forces.
Regarding the classes in the suggested order,
application of the aspect seems very debatable, even if some examples
may be found illustrating it.
Bases <Proteins < Carbohydrates < Lipids < Isoprenes<
kinones < porphyrines
- Bases< Proteins? (The inverse in histones !) How in rRNA,
in repressors?
- Proteins < Carbohydrates ? Examples could be Glycoproteins
(nearly all proteins in plasma are said to be of this kind,
and often proteins extending through cell membranes).
- Carbohydrates < Lipids ? Glycerine < fatty aids, okay.
The inverse in membranes? In glycolipids and gangliosides*
(much of these in grey areas of brains), the complex combination
of carbohydrates and fatty acids (including a transformation of
amino acid Serin), it may really be discussed which part binds
which.
- Lipids < Isoprenes as polyprenoles, okay, but
- Isoprenes < karotenes (fat-soluble) or quinones ?
- Quinones < porphyrines ?
* A ganglioside looks like an outline of an insect
larva
(fatty acid part) eating leaves (the carbohydrate hexoses).
In short, the more complicated molecules seem integrating several
steps along the suggested chain for classes.
d. The different classes of substances may be seen as undergoing
different number of steps from chains towards higher d-degrees on
superposed levels:
(5)-4-3-2-1: 4-5
levels of storage for DNA.
(4)-3-2-1: 3-4
for proteins as fibres, folded surfaces, globular forms to
enzymes
with coenzymes as 4-dimensional.
3-2-1 2-3
for carbohydrates (C6), chains to rings to macro-chains,
branched
or folded or spiralled to volumes.
2-1:
1-2 for steroids from isoprene chains.
9. Can the codon bases be connected with different classes of
substances?
Only to a certain extent as it seems. Here bases as nucleotides
- coenzymes.
According to limited data:
- TTP - cellulose synthesis
- UTP - carbohydrates, glucose synthesis, (also cellulose)
- CTP - lipids: connection between the amino acid Ser
and fatty acids
- GTP - participation at protein synthesis on rRNA (and
in the citrate cycle)
- ATP - general energy storing and transportations.
Most obvious is the connection UTP/TTP with carbohydrates. T/U-bases
represent directions inwards (T) towards DNA and outward direction
(U) towards active RNA, an essential opposition. (We could see the
similar opposition expressed in cellulose as inward" enclosing
cover in plant cells and the other carbohydrates inside the cells
or parts of them outward directed from lipid membranes of the cells.)
There is also the fact that most of the essential
amino acids which human beings cannot synthesize are U-base-coded.
All amino acids with U in 1st and/or 2nd position
in their codons derive from stations in the glycolysis of carbohydrates
(glucose →fructose), a fact that
presumably is connected with the role of UTP.
GTP-GDP keeps citrate cycle going around, by transforming Succinyl~
to Succinate. (Keto-acids in the cycle closely related amination.)
GTP-GDP is also a factor in the protein synthesis
at rRNA.
"G proteins" act as "molecular
switches", in transport of signals.
These data may give reason for connecting G-base more specifically
to proteins?
CTP is said to be more specific: taking part as a coenzyme in the
synthesis of glycerophospholipids and glycosylation of proteins
(Wikipedia.) It is involved in adding the amino acid Serin
to phospholipids.
ATP (with the A-base part in NADP, NAD and FAD) is obviously the
least specified, a main energy storing molecule and engaged in a
lot of processes.
AAA+ proteins: - many polarizing functions -
(as "protein degradation") and "intracellular transportations"
and motions in cilia...
As NAD the A-base is also involved in the respiration
cycle with ubiquinone (located to d-degree 1-- 0/00).
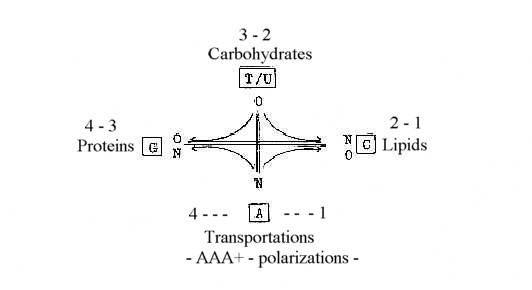
A summary could look like this:
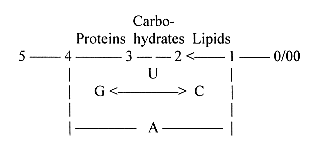
With the view on carbohydrates as fructose polarized a) towards
keto- and amino acids in synthesizing direction inwards and b) toward
fatty acids and lipids outwards, we get following picture, with
d-degree steps marked:
Three number operations - without sense?
Mass sums of bases with +1 for bond to ribose:
G 151, A 135, T 126, U 112, C 111.
Wiith U/T-bases regarded as representing the division of directions
of d-degree 4 (inwards: T/outwards: U), as in the views on protein
synthesis:
a) T/4 + A/1 + G/3 + C/2 = 1/2 x 544,666. 544 an essential
number in the mass analysis of the
genetic code.
b) 4 x U +3 x G + 2 x C + 1 x A = 1258, = sum of side-chains
of the 20 amino acids in the genetic
code, + 1 C + 1 A = + 246 = 1504 = 20 + 4 double-coded
ams R.
c. Dimension loops 4-1, 3-2 with similar numbers 141, 232, sum
373 =
mass numbers of T+U+A (126 + 112 + 135) and C+G+C ( 111 + 151 +
111).
373 in number-base system (nb-x) 8 = 251 in nb-10 =1/3 of 753, sum
of triplets in the elementary number series 5 - 0, 543 + 210 or
432 + 321.
*
To
- Fatty acids - some
general aspects
- 1/7 - fatty acids
and collagen
- Carbohydrates
|