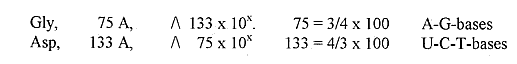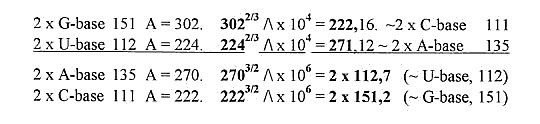RNA/DNA - Synthesis of the Bases:
The DNA-RNA-bases G and A versus C-U-T do indeed represent complementary
"poles", with several polarities in origin and their synthesis.
The 6-5-member-rings of G- an A-bases originate from Inosinic acid,
the 6-member-rings of U-C-T-bases from Orotate.
In original papers G and A are called "00"-bases, U-C-T
"0"-bases. Yet the G- and A-bases in their construction
have the character of 0-poles of the dimension model here, U-C-T
the character of 00-poles (anticentre poles).
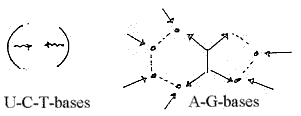
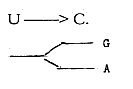
a) G- and A-base are constructed from a centre outwards, the U-C-T-bases
inwards:
The centre in G- and A-bases is made up of the smallest amino acid
Gly, whose branches outwards are filled with small molecules from
the surrounding anticentre.
We have a "c - ac"-relation .
The U-C-T-bases are created through the meeting from outwards of
two bigger molecules, the amino acid Asp and carbamyl phosphate,
binding to form the 6-member-ring.
Principle of synthesis for the opposite codon bases:
b) G and A then are formed along a branched way from
the resulting Inosinic acid, while U,-C and T are differentiated
along a "linear" path of changes.
c) The relation to the P-ribose groups of the nucleotides are opposite:
construction of A- and G-bases occur on the P-ribose group which
comes comes first,
U-C-T bases are constructed separately, with P-ribose groups are
added afterwards.
d) Gly and Asp, the amino acids making up parts of the polar types
of bases have inverted mass numbers:
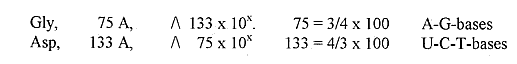
e) Mass number sums of "0"- and "00"-bass are
inversions:
G + A = 286 = 151 + 135 A (with
+1 for bond to ribose)
U+C+T = 349 = 112 + 111 + 126 A "
286/\ 349 x 10x
Perhaps we could note too, that the two parts in the synthesis
of U-C-T-bases,
(carbamylphosphatate + Asp) before binding has the mass sum 286,
the sum of A- and G-bases.
Also,more intricate:
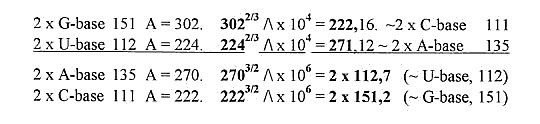
2 bases with exponents give their opposites if inverted.
The branched paths of synthesis for G- and A-bases:
Here we have something like a secondary polarity:
A-base gets its free N-group from the amino acid Asp,
the N-group in the B-chain of this. (There is of course no other.)
G-base gets its free N-group from the amino acid Gln,
the N-group in the R-chain of this.
We can note too:
Asp, 70 Z -------> A-base 70 Z.
Gln, 78 Z -------> G-base 78 Z
Codons as a mirror relation:
Asp G-A-C <------> C-A-G = Gln
but G-A-U <------> U-A-G = one of the Stop codons.
We could also observe the mass numbers of Asp, 133 A, Gln,
146 A and the sum of Orotate and Inosinic acid = 136 + 156
= 292 A: all numbers from the Exponent series: 52/3,
42/3, 32/3, 22/3(all x 100) etc.
(If written "5", "4" etc.: "5" = 292,
1/2 x "5" = 146, "5-2" = 133.
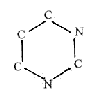
Forms of bases and amino acids:
There is a relation in their forms like centres-anticentres, (c-ac),
as 0-00 in the model - or radial versus circular as geometrical
poles of dimension degree 3:
- Amino acids as tetrahedrons have central C-atoms and direction
outwards in 3-4- directions,
- Bases are rings as if the central atoms had got inverted to anticentre
positions.
Formally, 2 amino acids N-C-C-N-C-C could be restructured to the
6-rings of the bases (if chemically possible is another question):
 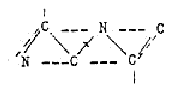
Some more elementary aspects on the 5 bases:

Elementary groups in the protein synthesis
- illustrated by the dimension chain and its superposed level:
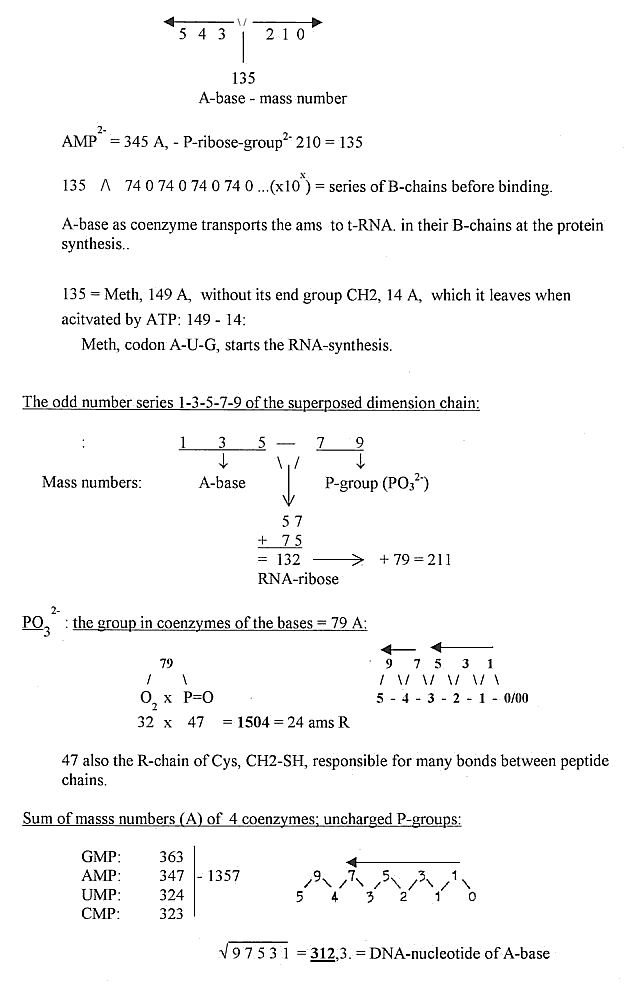
Numbers of DNA-RNA bases and some relations to
numbers of amino acids:
more here.
*
HOME
|