Codon bases, nucleotides:
From longer file on homepage here.
Used abbreviations:
Ams = amino acids, ama = single amino acid.)
The different side chains of ams = R-chains,
the common part, which bind through condensation: B-chains.
- Here is calculated with 24 ams, the four double coded included,
where nothing else is mentioned. See table over ams here.
/\ , sign indicating inverting
operation.
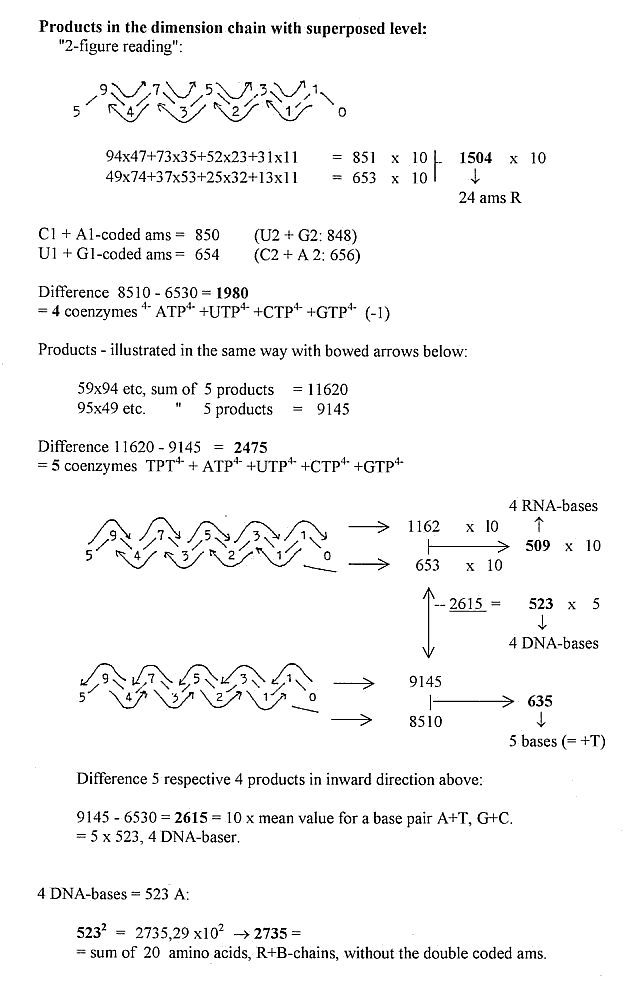
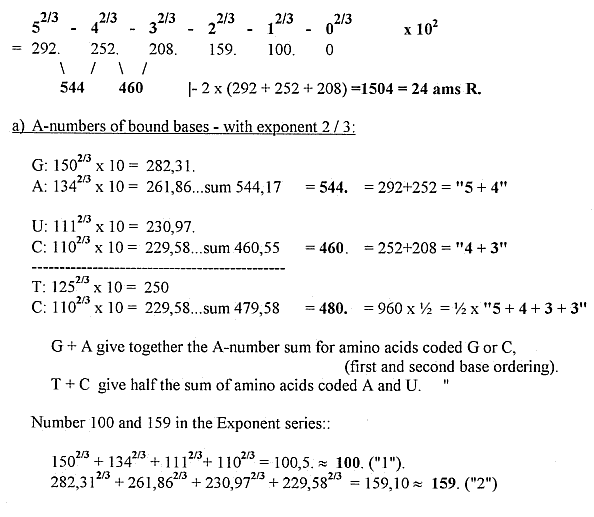
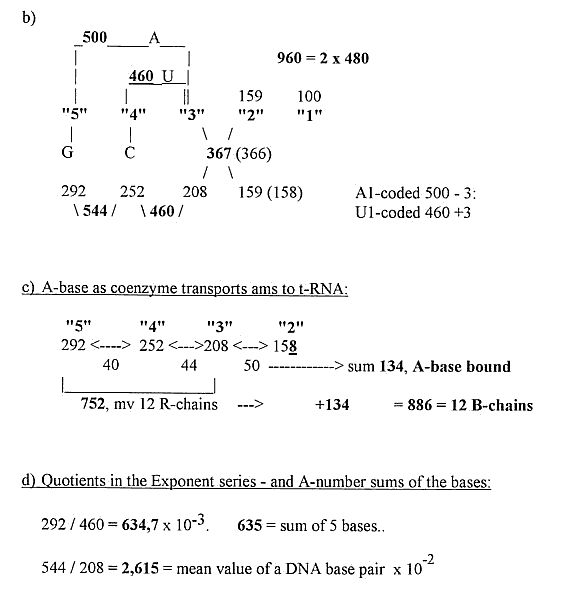
1) Total sums of amino acids (R) and 5-4 codon bases out of the
dimension chain:
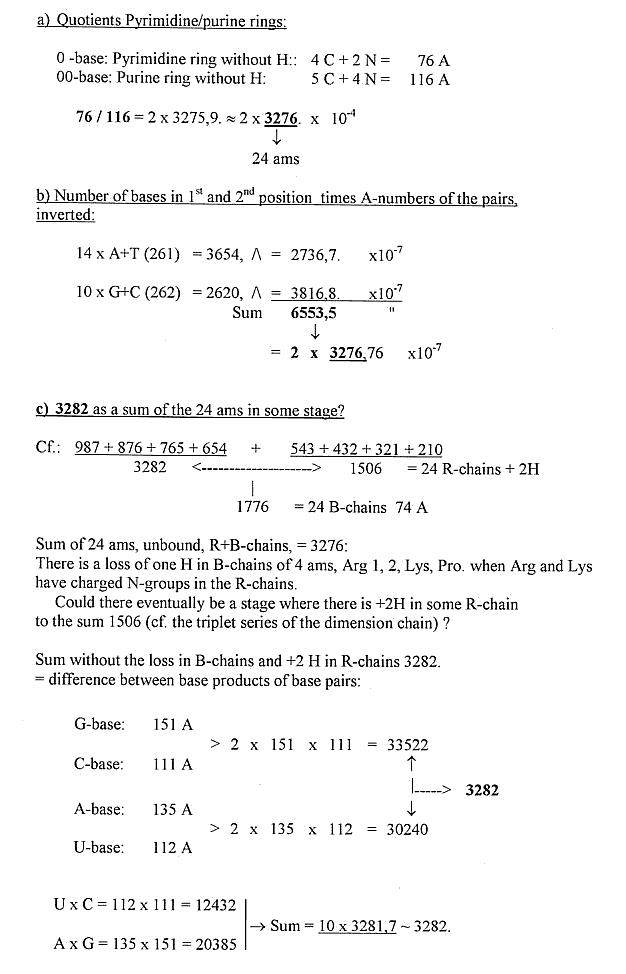
2) Some separate mass number relations Bases - Amino acids:
3) Triplets of nucleotides bound together
by only 2 P-groups:
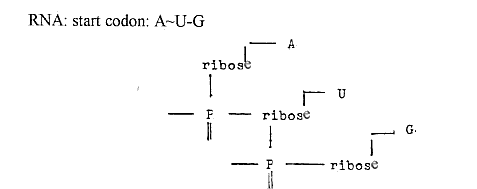
P-groups
ribose groups bases
bound
126 A + 2H 394
A 395
A
Nucleosides in bonds: 2 x 395, -1 = 790 -1 =
792 - 3
Corresponding nucleosides of the DNA-matrix, T-A-C-codon
= 714 +1
Triplets of a dimension chain:
Outwards Inwards
543 345
432 234
321 123
+ 210 +
012
1506 <-----> 714 . 1506
-2H = 24 amino acids, R-chains
|
792,
the interval ~ A-U-G-nucleosides
714 ~ T-A-C-nucleosides:
T-A-C--bases, bound = 369 A = 012+123+234.
Ribose part of the DNA-matrix: 3 x 116, - 2 H = 345 + 1.
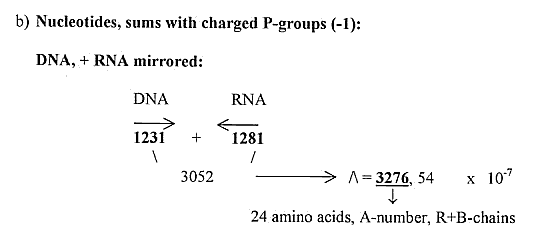
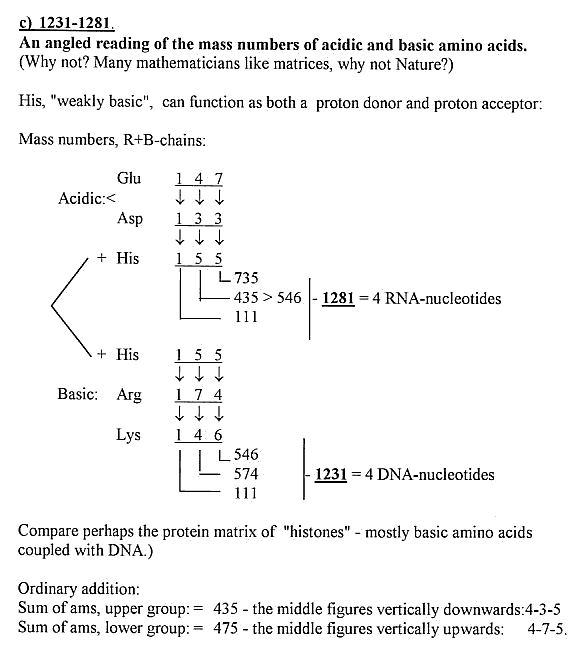
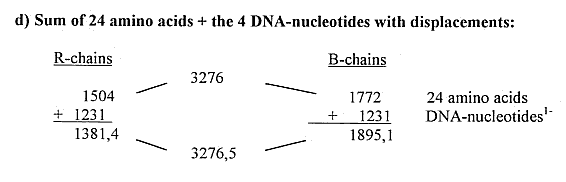
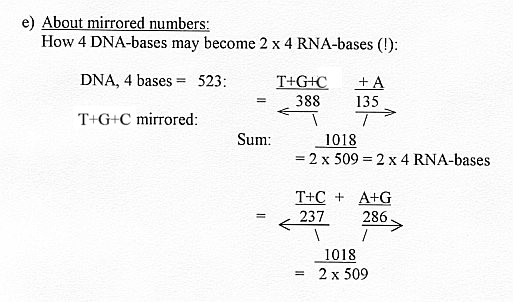
4) A variation of a "dimension chain" with 1-dimensional
"poles" in each step:
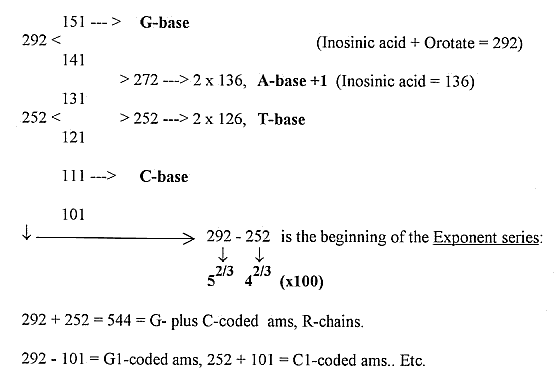
5) The "Exponent series" - the whole chain:
About the Exponent series,
see here.
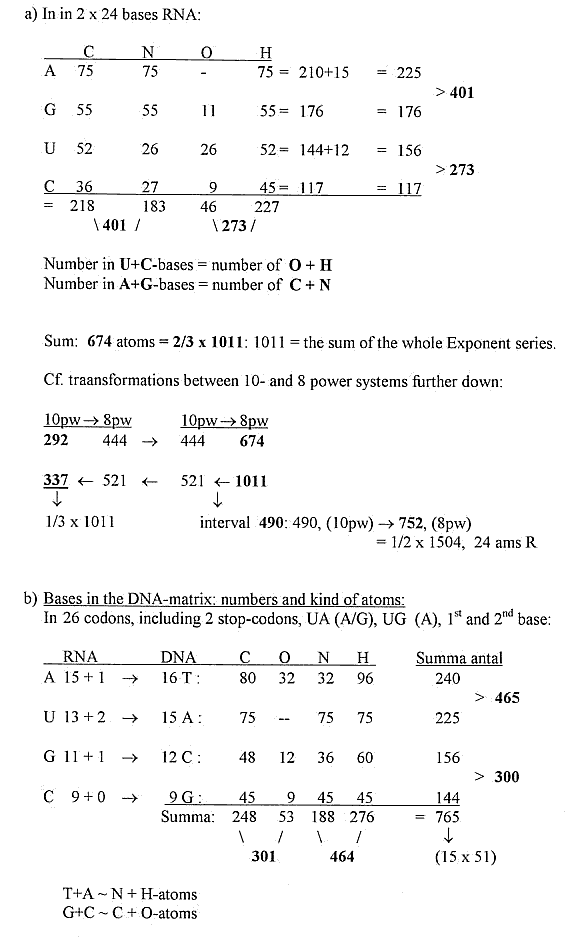
6) Number of atoms in codon bases in 1st and 2nd position
of the codons:
a) In in 2 x 24 bases RNA
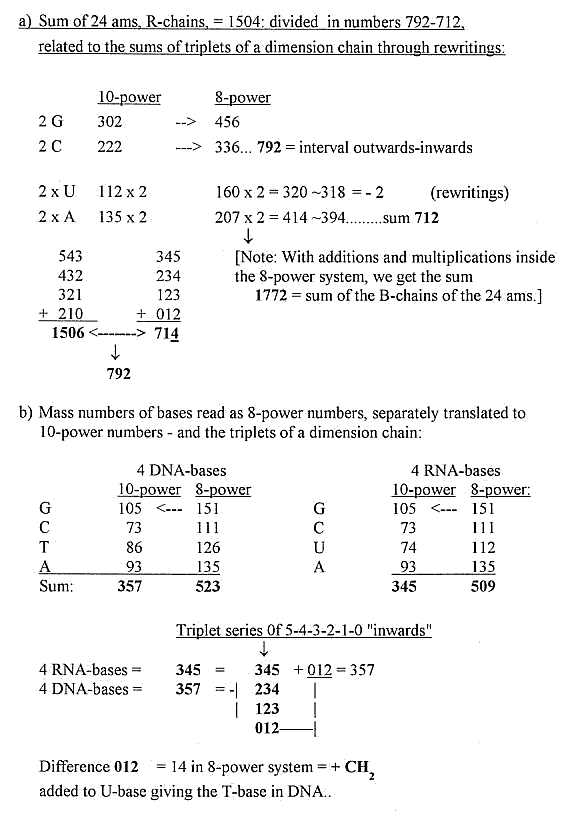
7) Transformations between 10- and 8-power numbers:
See further here.
Mathematical operations, i.e. additions, of 8-power numbers here
made
as if they appeared as 10-power numbers.
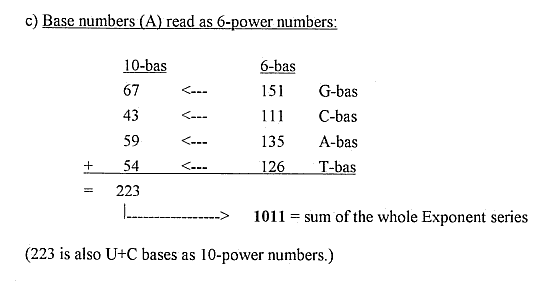
8) Free end groups of the 4 RNA-bases and the end groups
of amino acids
as Gln and Asn: A and T as a polarization step from G-, C-bases:
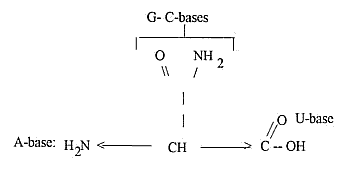
9) Serine and dimension chain as ~1/7 - see about Fatty
acids...
1 / 7: the inverted number 7
from triplets of a dimension chain as exponents of number 2:
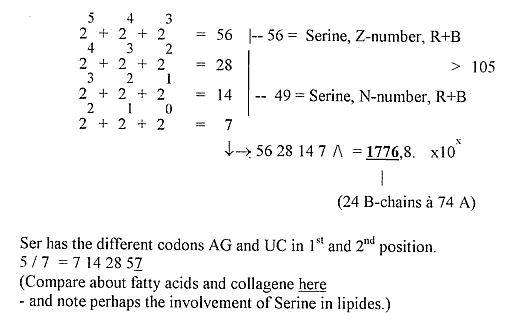
10) Hex-power transformations:
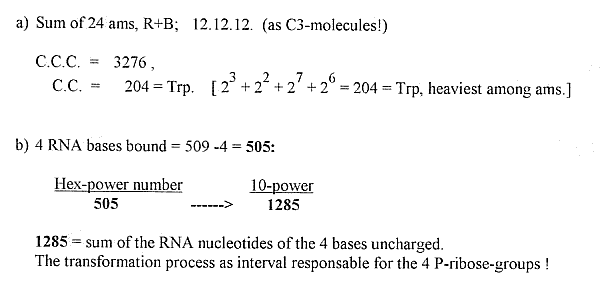
|















