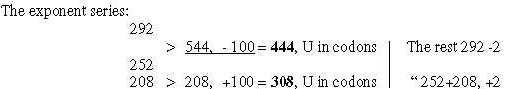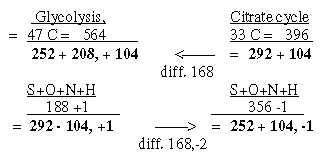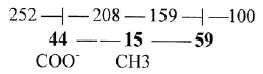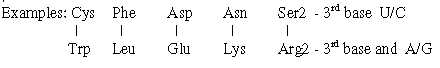1. Sum of ams with U-base in 1st and/or 2nd position of codons
is 752, exactly half the total = 292 + 252 + 208 in the ES-series.
It's division between the two 12-groups 770 and 734:
Group 770: Cross- and Form-coded: 208 + 100 = 308
Group 734: RNA- and Pair-coded: 544 - 100
= 444
All these ams derive from stations in glycolysis, no one from citrate
cycle.
(It's suggested here that this is connected with
the fact that U as coenzyme UTP-UMP is the one engaged in bonds and breaking of glycogen.)
Hence, we could imagine that the two groups or number chains à
752 also represent the two sides outside/inside mitochondria
membranes. This under following conditions:
Ala is regarded as derived from oxaloacetate (may
also derive from pyruvate). Ser2, AG-coded, likewise, as along the
outer loop from oxaloacetate via homoserine back to 3-P-glycerate.
Gly (one unit) from Ser1 or 2? Meth as connected with Cys (?).
With these assumptions we get ams derived from
the different stations in the processes as shown in table 6: Note
the approximate ES-number divisions
Table 6: Amino
acids from stations in glycolysis - citrate cycle: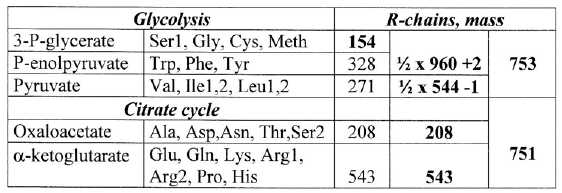
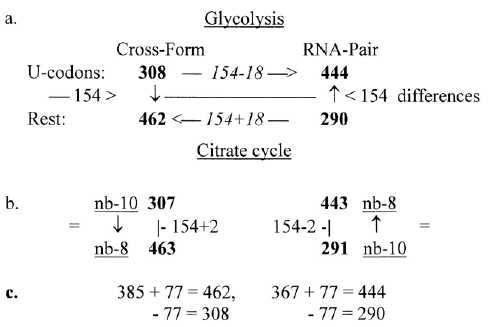
2. The divisions in codon type groups 770 and 734 on the both sides
of mitochondria membrane is shown in figure 9-1 below. Included
in the figure is an observation from files about number-base systems
that these groups -/+1 seem to refer to each other through transformations
between nb-10 and nb-8 and in opposite directions:
Fig 9-1: Codon
type groups, division on Glycolysis and Citrate cycle:
(The codon type groups defined in file 2.)
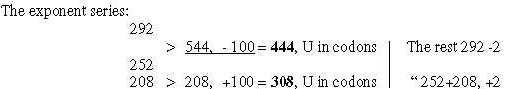
Differences, cf. interpretation of number 77:
308 - 444 = 136 = 4 x 34
462 - 290 = 172 = 4 x 43 (Cf. A1 - U1 = 1 x 34, A2 - U2 = 2 x 43)
43 to 34: 9 ~ OH- Z
Also the rest, not U-contenting codon groups, are divided +/-2
in numbers of the ES-chain, a division in step 5 - 4, while U-groups
represent a number division in step 4 - 3.
In the figure above we have for instance 18 as differences 308-290,
444-462 and +/-2 in the transformations. We have perhaps (?) here
a correlation among ams to the general theme in the processes, secretion
of 2H and processing of H2O (18). If
so, what about +/- 154? Could it eventually express
the same as the loop, outside the main steps, from oxaloacetate
via homoserine (and homocysteine) back to 3-P-glycerate where sum
of ams = 154?
Codons and ams in the four groups are shown below:
Fig 9-2:
Amino acids in the 4 codon type groups:
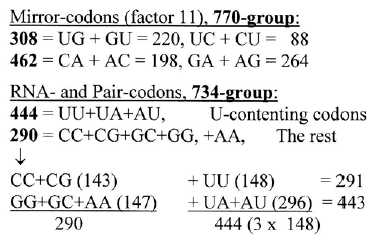
3. Division in G+C- and A+U-coded ams between the two parts of
the process shows notable agreement with divisions in the ES-chain
(+/-1).
Fig 9-3:
Division on groups G+C, A + U from stations in Glycolysis - Citrate
cycle:
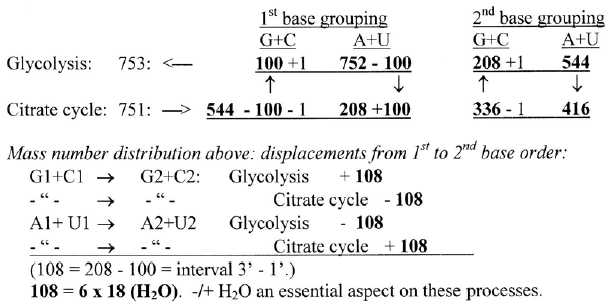
4. Atom divisions on ams from Glycolysis and from
Citrate cycle become the same for C-atoms as between U1+C1-coded
and G1+A1-coded ams (cf. file 4, figure 4-1):
Glycolysis 47 C = 564 = 460 + 104, other atoms 292 - 104 +1.
Citrate cycle 33 C = 396 = 292 + 104, other atoms 252 + 104 -1.
Fig. 9-4: Division on atom kinds
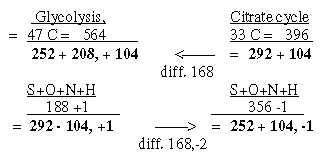
5. Sums of stations in the two processes, compared with groups
of ams, a) bound, b) with +1 for charge and bonds to S- or P-enzymes:
Glycolysis, 8 C3-stations: a) 736, b) 750.
Citrate cycle, 10 stations*: a) 2 x 736, b) 2 x 748
*Malate, Oxaloacetate, Citrate, Cisaconitate,
Isocitrate, Oxalosuccinate, α-ketoglutarate,
Succinyl(~Coa), Succinate, Fumarate.
That the sums approximate numbers as 752 and 734 among ams in the
ES-series is not so natural as it perhaps may seem.
Table 7: Molecules in the citrate cycle
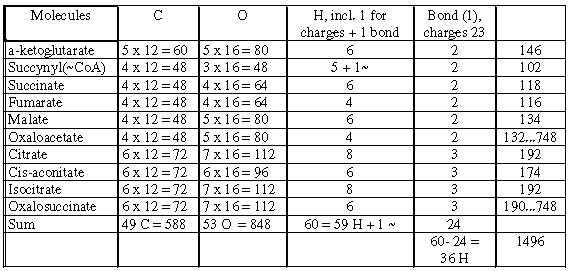
1496 = 11 x 136 (~ Hypoxanthine) = 44 x 34.
Molecules with 5 C and 4 C = 748 - 12 (6 x 2) for charges, bond = 736;
molecules with 6 C =:748, - 12 (4 x 3) = 736. .
736 = 32 x 23.
Sum of substances
= 1496, x 2 (turns) = 2992 = 11 x 272 (half the number 544)
Eight of the stations in Citrate cycle as uncharged,
compared with the ES-chain:
101 = Succinyl →
→ 2 x 59 = Succinate →
→ → 2
x 58 = Fumarate →
→ → → 2 x 133 +/-1 (== 292 - 159)
Malate + Oxaloacetate →
→ → → → 2 x 192 (292 - 100) = Citrate + Isocitrate →
→ → → → → 146
(½ x 292) = α-ketoglutarate, → (→
Succinyl...101)
(Two substances excluded here, Cisaconitate 174 and Oxalosuccinate 190.)
6. 44 - 59, intervals in the E-chain:
It is worth noting the intervals 44 (4' →>3')
and 59 (2' ← 1')
in the ES-chain, steps that correspond to each other in the background model. See figure 9-4. It is numbers equivalent with main molecules incorporated into the citrate
cycle: CO2 in step from Pyruvate (88)
to Malate (133+1) and of acetyl-(CoA) + O(H) = 59 (60) in step from
Oxaloacetate (133 -1) to Citrate 192 (132 + 60 (~ 133 -1 and 59 +1):
Fig 9-5: Intervals
44 and 59 in ES-chain as additions into Citrate cycle:
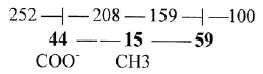
7.
Orotate <———> Hypoxanthine; codons U/C → A/G in 3rd position:
There is a trend from ams with 3rd base U/C to ams with 3rd base A/G like a coordinate axis through the processes, in ams codons from first stations in the glycolysis to Glu in the citrate cycle, illustrated by following examples:
Fig 9-6: Orotate - Hypoxanthine, a coordinate axes through the processes
8. Anti-codons:
Most codons of ams from Glycolysis have anti-codons that are codons
for ams from the Citrate cycle - as a kind of "complementary
poles". Is it perhaps possible to imagine that the two series
of codons once originated from opposite strands of DNA (via tRNAs)?
One such variant is shown in table 7:
Table 8: Anti-codons:
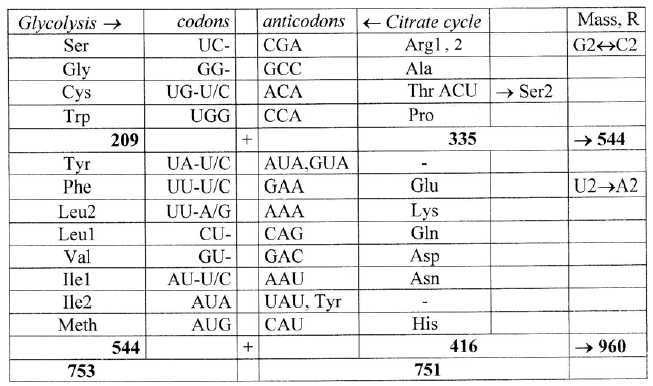
Among ams from Citrate cycle the AG-codons of double-coded Ser
and Arg can only be derived in a second step through a change between
optional U and C in 3rd base as from Thr and Pro.
Exchanges within group 416 in the table are possible
*
To The bases
- some annotations.