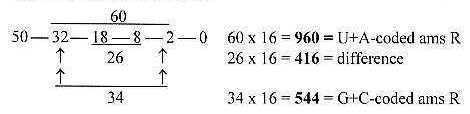1. Mass division on kinds of atoms:
First to observe is the fact the mass division on C-atoms versus
other atoms is the same as between U+A-coded ams and G+C-coded ones:
C-atoms, 80 = 960
N + O + S + H = 544
The division on codon grouped ams of the "substituents", N, O, S, H = 544:
G1 + A1: 292
C1 + U1: 252
Mass of C-atoms in 12-group 770 = 444 = 544 - 100
Mass of C-atoms in 12-group 734 = 516 = 416 + 100,
H-atoms: 252 - 100 = 152 = 4' - 1'
N+O+S-atoms: 292 + 100 = 392 = 5' + 1'
The division between N-atoms and S+O+H-atoms could perhaps be noted
too:
Mass of atoms in R: C + N = 3 x 376, ½ (5' + 4' + 3'). O + S + H = 376
10 O + 2 S + 152 H = 376 = 1/2 x 752
|- 2 x 208
12 N = 168 = 1/2 x 336 (544 - 208).
80 C + 12 N = 1128 = 3 x 376
2. The division of C-atoms on codon groups Purine - Pyrimidine pairs:
and simultaneously on codon groups: s
G1 + A1: 33 C = 292 + ½ x 208 = 396
C1 + U1: 47 C = 460 + ½ x 208 = 564
When one of the two 208-groups is divided on first numbers 292
- 252, we get the relations between mass division on atom kinds
and on codon groups of ams shown below:
Fig 4-1: Division
of atom kinds and ams groups, halving 208
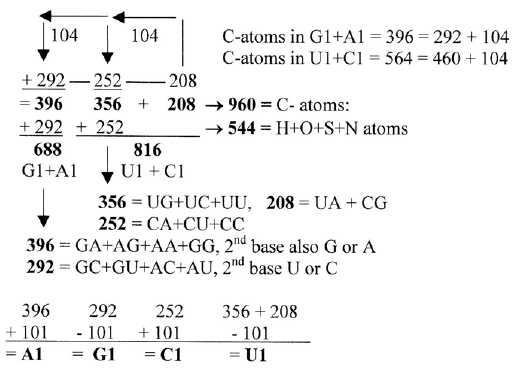
In table 4 below we find number 104 as the difference between groups
G1 +C1 and U1 + A1. With number 416 uncounted, the sums get equally
divided vertically and horizontally.
The arrangement could perhaps reveal a symmetric
phase in step 5'- 4' along two axes (544) in opposite directions:
C-atoms to/from "substitutes", before a stage 3'
(416).
3. The keto-/amino acid polarity:
Table 4: Mass
of C-atoms in G+C = mass of other atoms in U+A
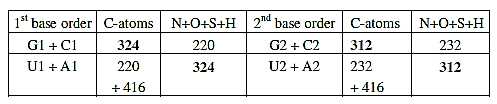
The domain groups of ams along the third polarity between
bases, the keto-/amino types, shows in 2nd base order a mass division
on atom kinds with strong regularity:
C-atoms: G2 + U2 = 576 = 3 x 192 (292 - 100). Other atoms
= 272
C2
+ A2 = 384 = 2 x 192 (292 - 100). Other atoms = 272
Cf. in B-chains unbound: C = 576, O = 2 x 384; N = 336 (544 - 208),
H 96 - 4.
4. Number 192:
This number 192 appears in many sums in the genetic code. Why?
In the ES-chain = 5' - 1', the interval 292 -- 100.
(I's the mass of citrate, isocitrate, however not an explanation. It's 3 times 64, 6 times 2^5.).
1536 - 1344 - 1152 - 960 - 768 - 576 - 384 - 192
8 7 6 5 4 3 2 1 x 192
1536 = sum of all C-atoms in 24 ams, R + B.
1344 = sum of 24 bound B-chains (24 x 56)
960 = U+A-coded ams
768 +2 = ams with mixed codons, 2 x 384
576 -1 = U+A-group in non-mixed codons
192 -1 = G1-coded ams
5. C-atoms in R-chains
of ams as basis for mass division:
This division doesn't concern codon distribution but seems related
to the ES-series and number 584, 2 x 292, with certain assumptions.
(Here C for carbon.)
Phe and Tyr are synthesized as 3C- plus 4C-molecules,
Trp as 3C + 4C + 5C - 1C. Trp gets its B-chain from Ser, shares
codon with Cys and can brake down to Ala, hence here regarded as
"meeting the other way around", added to the l C group.
Fig 4-2:
Cn: 4C, 7C, 3C +0C, 2C, 1C + 9C

(4 C: Arg1, Arg2, Lys, His = 356, + Leu1, Leu2, Ile1, Ile2 = 228.
7 C: Phe + Tyr = 198;
3 C: Glu, Gln, Val, Meth, Pro = 305; 0 C =
Gly = 1;
2 C = Asp, Asn, Thr = 162;
1 C: Ala, Ser1, Ser 2, Cys = 124;
9 C:
Trp = 130.)
6. Number of C-atoms in codon groups of ams as halvings:
We may also note that number of C-atoms are approximately distributed
on codon groups as 2/3 - 1/3 in two steps
53 C in U1+A1-coded ams (+1 in A2 + U2), ~ 2/3
x 81
27 C in G1+C1-coded ams (- 1 in G2 + C2),
~ 1/3 x 81
↓→18 in C1, 19 in G2, ~ 2/3 x 27
9
in G1, 7 in C2, ~ 1/3 x 27
7. End atoms of R-chains
as basis for mass division:
- Ams with end atoms CHx plus Gly (H) = 420.
- Ams with end atoms O, OH, S and no N = sum 468
- N in end-groups or in rings (Trp and His), including Gln and Asn
= 616.
420 + 468 = 2 (544 - 100) = 888
616 =
2 (208 + 100).= 616*
*(2 Arg charged = 200 +2, rest 416 - 2)
Note that all N-contenting ams but Trp derive from the citrate
cycle.
(Another division: ams more or less polar: 468 + 616 = 2 x 544 -
4. Non polar ams, the CHx-group = 2 x 208 + 4.)
7x: Three notes:
a) A division of the G+C group of aa 544, 1st base order, after kind of end atoms, approximates the one between mixed and non-mixed codons, 385 and 159:
b) [ U1 +A1 group 960, if S-contenting aa are added to the CHx-group:
CHx: UU, UU, AU1, AU2, =262
S UG, AUG = 122....................................................Sum 384, 2 x 192
O: UA, UC, AC, AG = 214
N: UGG, AA, AG = 304.......Sum 518 (2 x 259)
O-N AA = 58.............................+ 58............. .........Sum 576, 3 x 192 ]
c) RNA- and Pair-coded aa divided after end atoms:
H + CHx: 7 aa 2 x 208 - 2
S, OH, O-N, NHx: 5 aa: 2 x 159 + 2
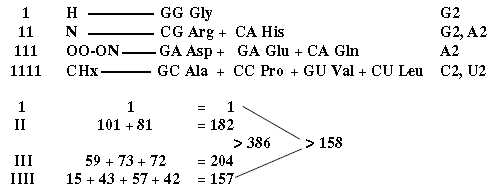
8. 81 - 47, a division of number 128 and number of
atoms:
There are 128 C-atoms in the R- + B-chains of the 24 ams 128 = 544 - 416.
His 81 and Cys
47 (mass of R-chains) cooperate in an enzyme to break fructose at the start of the glycolysis.
The number of atoms in R-chains are exactly twice the total
number of C-atoms in R+B-chains in groups G + C and U + A:
G1+C1: R+B = 47 C →
all atoms in R = 94; (G2 + C2 46 and 91)
U1+A1: R+B = 81 C → all atoms in R = 162; (U2 + A2 82 and 165)
Fig. 17 files, 03-03: 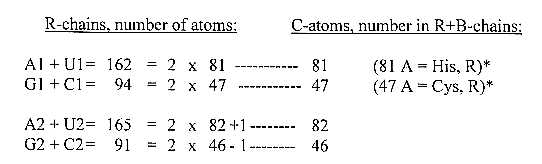
Which skeleton doubles its projection in its radical part? The
human body in the brain!?
U1 + A1-coded ams: 81 C = entire number of C-atoms
in R-chains +1
G1 + C1-coded ams: 47 = entire number of C-atoms
in B-chains - 1
Minus / plus 81 and 47 gives also from the codon groups of ams from 544 and 416:
½ x 544 - 81 = G1, 191
½ x 544 + 81 = C1, 353
544 - 81 = U1, 463. 544 - 47 = A1, 497
416 + 81 = A1, 497
416 + 47 = U1, 463....difference 34 = 81 - 47.
Why this division of 128 in 81 - 47?
Cf. figure 13-2 in file 13 about the 2x2-chain:. Total 94 x 16 or 47 x 32 = 1504
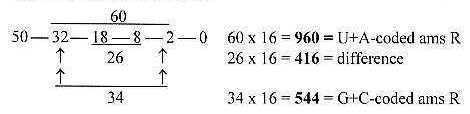
94 + 34 = 128, ½ x 94 = 47, + 34 = 81
Cf. R-chains of His 81 nd Cys 47 making up the active center of he enzyme halving fructose at the start of glycolysis.
9. A similar "perpendicular" relation between number of C and H
in the same codon groups:
C + H: total in U+A-coded ams R = 152, ~ number of H in all R-chains
C + H: total in G+C-coded ams R = 80, ~ number of C
in all R-chains.
(See figure here.)
10. Some gatthered points from above and other files
with a link to tables in an Appendix:
1. In ams groups G1 + C1 and U1 + A1 the number of atoms in side chains R is exactly 2 times the total number of C-atoms in R+B chains; (File 04 §8):
G + C: 47 C (R+B) → 94 atoms (R)
U + A: 81 C (R+B) → 162 atoms (R).
2. R + B chains unbound:
G1 + C1 differs from G2 + C2 with 1 unit: 1282 → 1281.
U1 + A1 1994 → U2 + A2 = 1995.
3. The keto-/amino acid polarity, ams R+B unbound, sum 3276:
A1 + C1 = G2 + U2 = 1808 Number of atoms also the same1 267
G1 + U1 = A2 + C2 = 1468 - " - : 201
1 However, not equally divided on atom kinds.
Cf. Transformations, file. 17, § 1.4.1.
4. The keto-/amino acid polarity, R-chains, (file 04-§3).
A2 + C2 —
G2 + U2: 384 — 576, a 2/3 division of C-atoms
544 divided equally on the groups: 272 — 272: 'substituents'.
5. Mass division between purines and pyrimidines: file 04 § 1, 2.
544 into 292 - 252 between purines and pyrimidines:
A1+G1: C-atoms 292 + 104 = 396. Substituents 292
U1 + C1: C-atoms 252 + 104, + 208 (= 564). Substituents 252
6. R+B bound, sum 2848:
C-atoms 1536, substituents 1312 = 4 x 192 (B) + 544 (R)
See file 13 §6, 1760 - 224-steps.
7. R+B bound:
Number of atoms without C = 272. (+ 128 C = 400.)
Condensation from 468 atoms, R+B unbound, implying + 4H - 3 x 24.
8. 20 ams: 384 atoms, the two sets of ams with two codons = + 84 atoms (5' - 3').
384 divided on R- and B-chains 207 — 177, about the same division as in the ES-chain and Table 1 on ams with mixed codons.
Link
to Appendix:
Tables on mass division on atom kinds and number of atoms, 1st
and 2nd base order, in R, R+B unbound and in R+B bound.
To The two 12-groups
of ams - details.