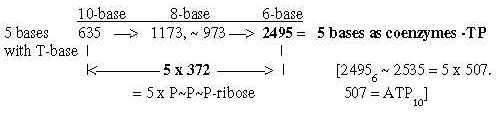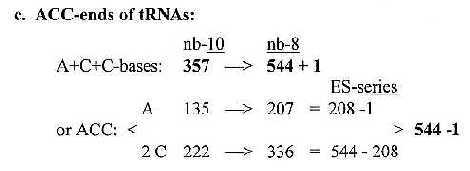|
|
|---|
|
Astronomy
- links:
|
Elements - links:
|
The Genetic Code -
links:
|
Language - links
|
|
P- phosphorous groups - Coenzymes - Nucleotides - Met AUG
|
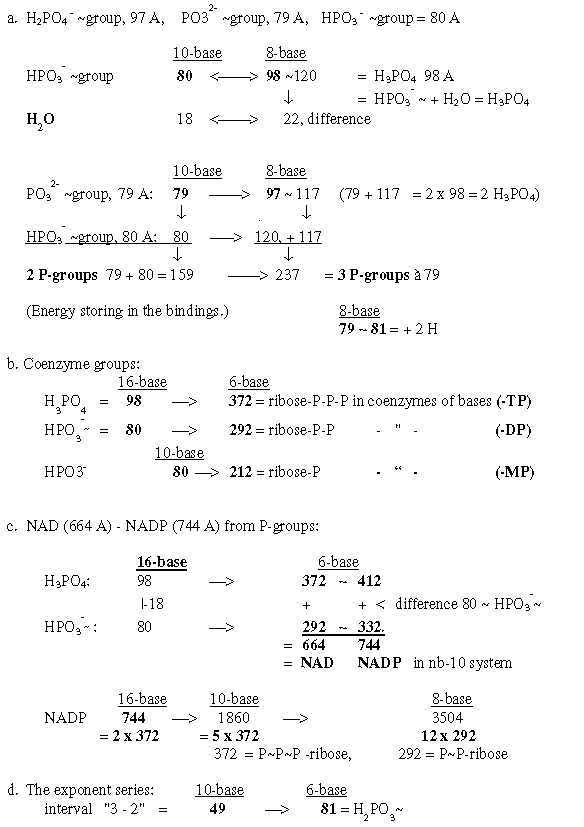
1. P-groups, the single, "inorganic" phosphorous groups: Fig 19-1:P-groups: A form of life was found some years ago, said to use arsenic instead of
phosphorus (P), i. e. next higher element in the phosphorus group of
elements in the periodic system. If so, it could of course lead
to the conclusion that all such transformations between masses including
phosphorus are irrelevant and in any case no necessary condition
for life as an eventual part of a reference system.
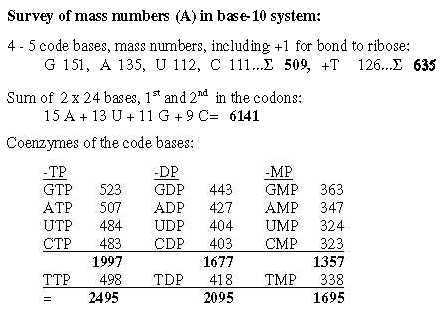
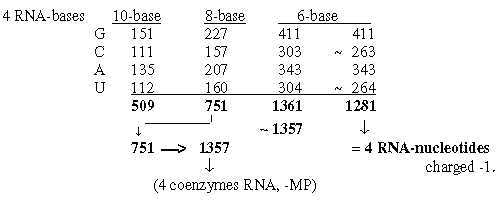
2. Coenzymes of the bases, -MP, -DP, -TP: Fig. 19-2:Survey 2.2 From 4 bases to their mass as coenzymes Fig 19-3: 509 - 1357, 4 coenzymes -MP:
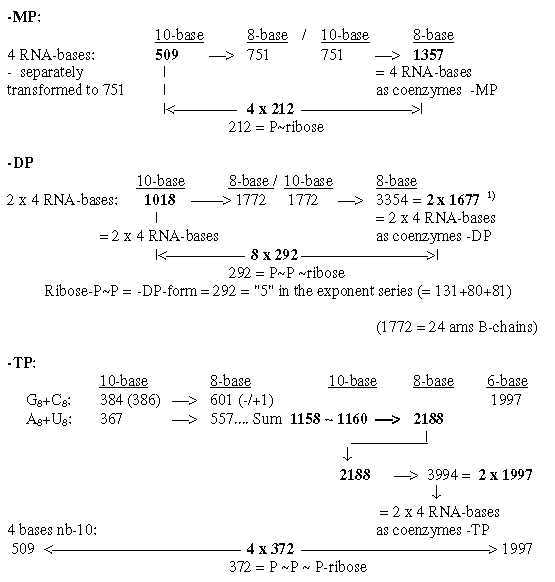
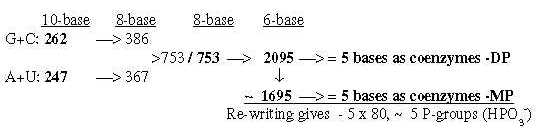
2.3 Expansion of bases nb-10 to nb-8 adds the Px-ribose groups: Some transformations from sums of the bases to sums of their appearance as coenzymes are shown in figures below. Note expansions where 212-292-372 correspond to the P(P(P)-ribose groups: Fig. 19-4:From the bases to coenzymes -MP, -DP, -TP
Fig. 19-6: 2.6 From 751, the sum of 4 bases in nb-8, Fig. 19-7:
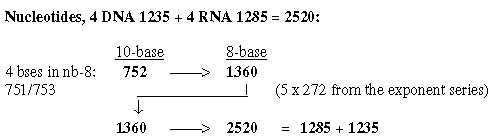
3. Nucleotides: 3.1 Survey of nucleotides in chain binding: Fig 19-8:
Fig 19-9:
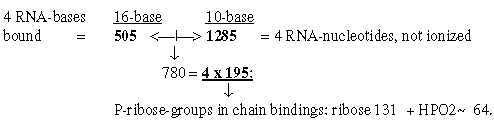
Fig. 19-10:
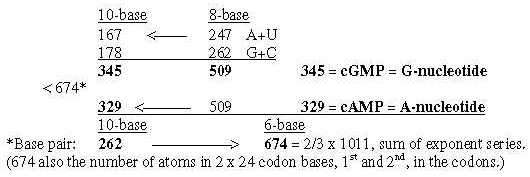
Fig. 19-11: cGMP - cAMP:
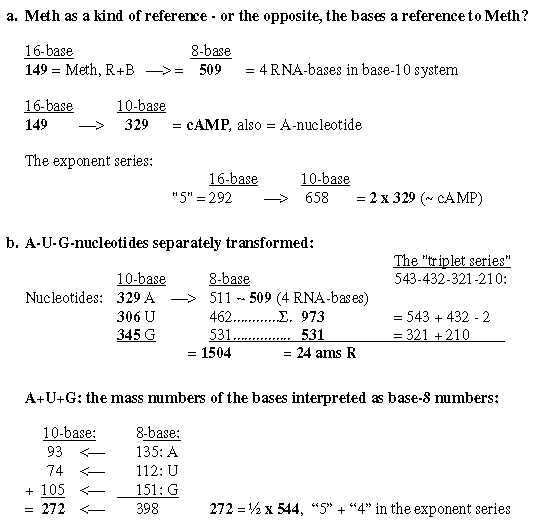
4. Met - codon AUG and tRNA-ends ACC: Fig 19-12: AUG, codon for Meth:
5. A-C-C - ends of tRNA: Fig 19-13: tRNA-ends ACC:
Cf. mass numbers for A and C from Triplets, file 21; 012 + 123 = 135 (A-base), + 234 = 357. Two of the intervals in the steps = 2 x 111 (2 x C-base). *
|
|
|
© Åsa Wohlin
Individual research
E-mail: a.wohlin@u5d.net
Table
24 amino acids (ams)
R-chains, A, Z, N
Abbreviations
- ways of writing -
Background
model
Files here:
All
these files gathered
in one document, word,
124 p.
All these files in 3 documents, pdf:
Section
I, files 0-11
Section
II, files 12-16
Section
III, files 17-22
Discusssion, References in section III
To
17 short files.
- partly other material
-
The
17 files as one document,
pdf
An
earlier version (2007)
with more material
on the same subject, 73 pages:
Contact:
u5d
Latest updated:
2022-11-12